Figure 1.
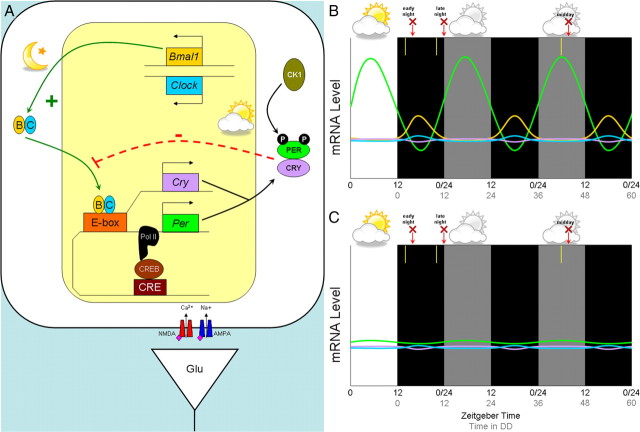
The molecular components of the circadian clock and their circadian expression in a retinorecipient neuron of the SCN. A, The circadian clock gene/protein network that drives an ∼24 h period contains a positive limb with BMAL1 and CLOCK activating transcription of Per and Cry. PER and CRY proteins are phosphorylated by casein kinase 1 (CK1) and form the negative limb by downregulating their own transcription. B, C, Typical expression of generalized circadian genes over time in a wild-type mouse (B) and in a Bmal1−/− mouse (C), based on data from Pfeffer et al. (2009). White and black backgrounds represent lights on (daytime) or lights off (night), while a gray background represents “subjective day” in constant darkness. The y-axis refers to relative mRNA levels, and the x-axis refers to time (in hours), with the black numbers corresponding to zeitgeber time (ZT; where ZT 12 refers to lights off) and the gray numbers corresponding to hours in constant darkness. In the study by Pfeffer et al. (2009), 15 min light pulses were given at 2 h after dusk (early night), 2 h before dawn (late night), or after 42 h in constant darkness (“subjective day”) as indicated by the yellow bars; animals were killed 2 h after each respective light pulse as indicated by the red arrow and “X.” Genes correspond by color to those introduced in A.