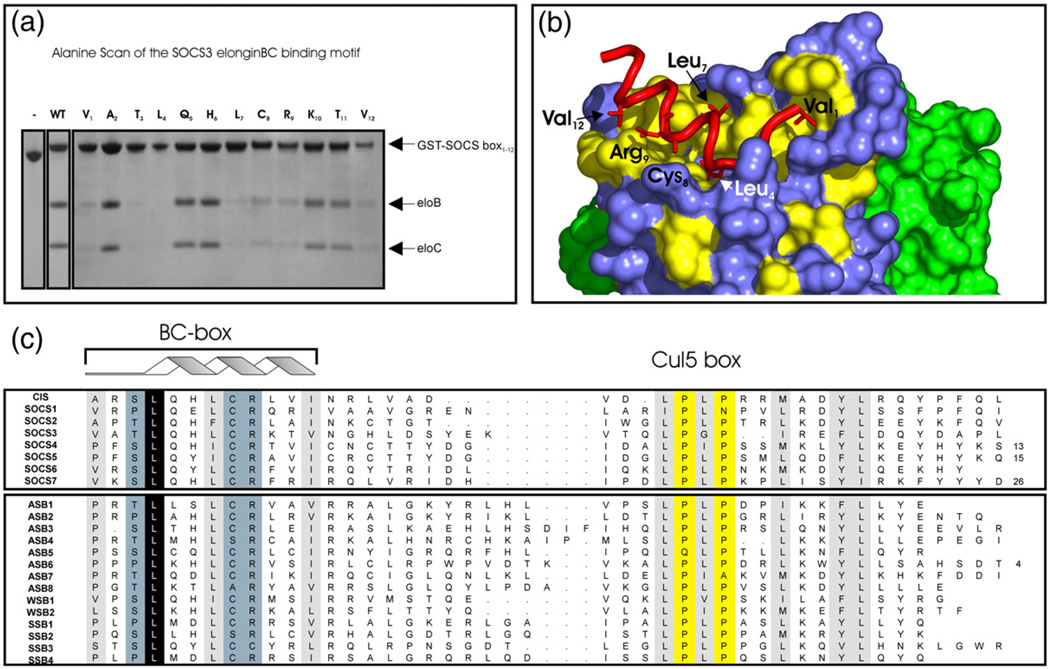
Fig. 6.
Key residues required for elonginBC binding. An Ala scan of the SOCS3 SOCS box to determine residues required for elonginBC binding was performed. Co-expression of 12 SOCS box domain constructs, each containing a single Ala mutation, with elonginBC was performed in E. coli, and glutathione Sepharose was used to pull down GST-labelled proteins present in the cell lysate. (a) The following mutations completely interfered with elonginBC binding: Val1, Thr3, Leu4, Leu7, Cys8, Arg9 and Val12. Of these, only the L4A mutation completely abolished binding. (c) Residues identified by Ala scan are highlighted on a surface representation of elonginBC where hydrophobic residues on the surface of elonginC are shown in yellow. The BC box of SOCS3 (red) is shown in cartoon representation with important side chains displayed in “stick” representation. ElonginC is shown in blue and elonginB in green.