Figure 2. Super-enhancers are associated with key ESC pluripotency genes.
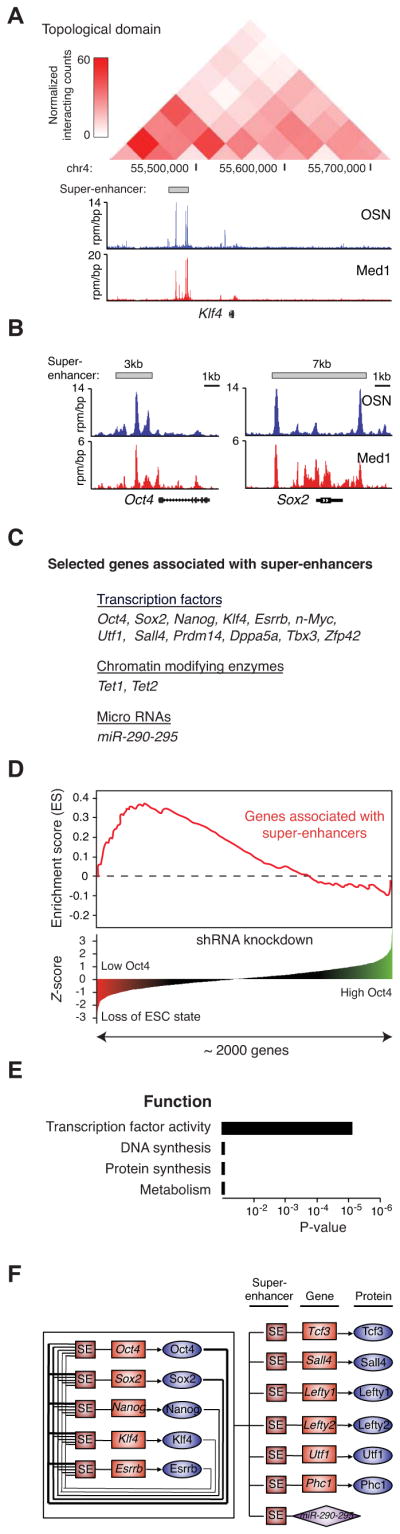
A) ChIP-Seq binding profiles for OSN and Med1 at the Klf4 locus in ESCs. Previously described Hi-C interaction frequency (normalized interaction counts), and genomic coordinates comprising a portion of a topological domain (Dixon et al., 2012), are indicated above the binding profiles.
B) ChIP-Seq binding profiles for OSN and Med1 at the Oct4 and Sox2 loci in ESCs. Gene models are depicted below the binding profiles.
C) List of selected genes associated with super-enhancers and playing prominent roles in ESC biology.
D) Super-enhancers are associated with genes encoding transcription factors, coactivators and chromatin regulators important for maintenance of ESC state. The gene set enrichment analysis (GSEA) of super-enhancer-associated genes reveals these genes encode regulators whose shRNA knockdown most impact Oct4 expression and ESC state. Knockdown of genes important for ESC identity causes Oct4 loss, which is reflected by a negative Z-score. P-value (10−2) was calculated as part of the GSEA analysis.
E) Gene ontology (GO) functional categories for super-enhancer-associated genes. Genes encoding factors important for DNA synthesis, protein synthesis, and metabolism were not enriched. See also Figure S2.
F) Schematic diagram of a small portion of the ESC core regulatory circuitry. Genes encoding the master transcription factors are themselves driven by super-enhancers. These master factors form an interconnected feedback loop and regulate their own expression, as well as the expression of other transcription factors and non-coding RNAs that play prominent roles in ESC biology.
