Figure 3. Super-enhancers confer high transcriptional activity and sensitivity to perturbation.
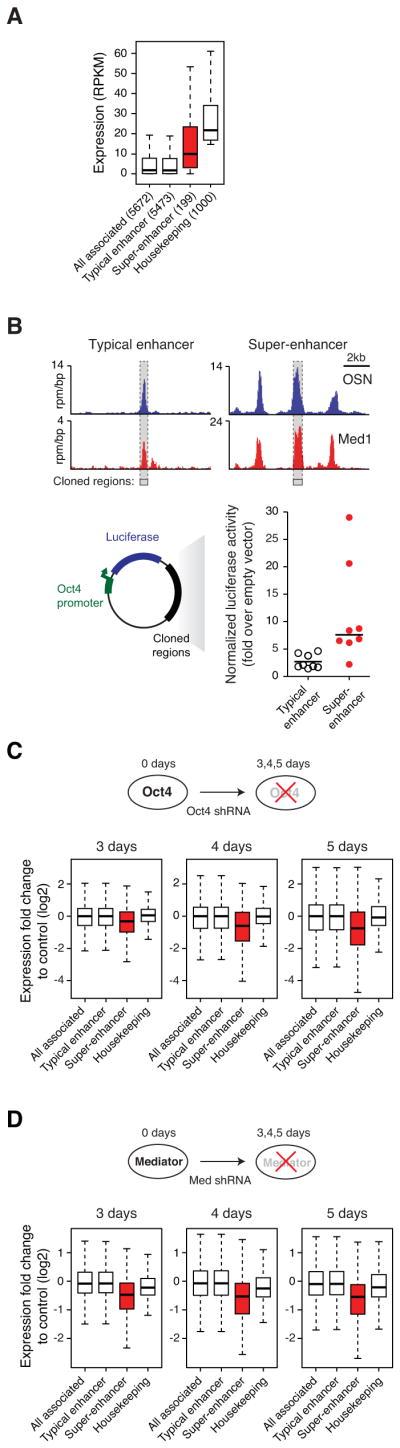
A) Box plots of expression (reads per kilobase of exon per million mapped reads (RPKM)) from typical enhancer-, super-enhancer-, and all enhancer-associated genes, as well as the top 1,000 highest expressed housekeeping genes. The number of genes belonging to each category for which we have expression data is denoted. P-value (10−5) was calculated using a two-tailed t-test. See also Figure S3A and Table S4.
B) Upper panel, ChIP-Seq binding profiles for OSN and Med1 at the Sgk1 typical enhancer and Esrrb super-enhancer loci. Grey bars with dashed borders indicates cloned regions. Lower panel, Plot of luciferase activity 24 hours post transfection, normalized to a transfected control plasmid. Enhancers neighboring selected genes were cloned into reporter plasmids containing the Luciferase gene regulated by the Oct4 promoter, and subsequently transfected into ESCs. P-value (0.02) was calculated using a two-tailed t-test. See also Figure S3B.
C) Upper panel, Schematic diagram of ESCs transduced with shRNAs against Oct4. Lower panel, Box plots of fold change expression in differentiating ESCs (3, 4 and 5 days after transduction) relative to control ESCs transduced with shRNAs against GFP. P-values (day 3= 10−5, day 4=10−8, day 5= 10−10) were calculated using a two-tailed t-test. See also Figure S3C.
D) Upper panel, Schematic diagram of ESCs transduced with shRNAs against the Mediator subunit Med12. Lower panel, Box plots of fold change expression in Mediator-depleted ESCs (3, 4, and 5 days after transduction) relative to control ESCs. P-values (day 3= 10−11, day 4=10−11, day 5= 10−13) were calculated using a two-tailed t-test.
