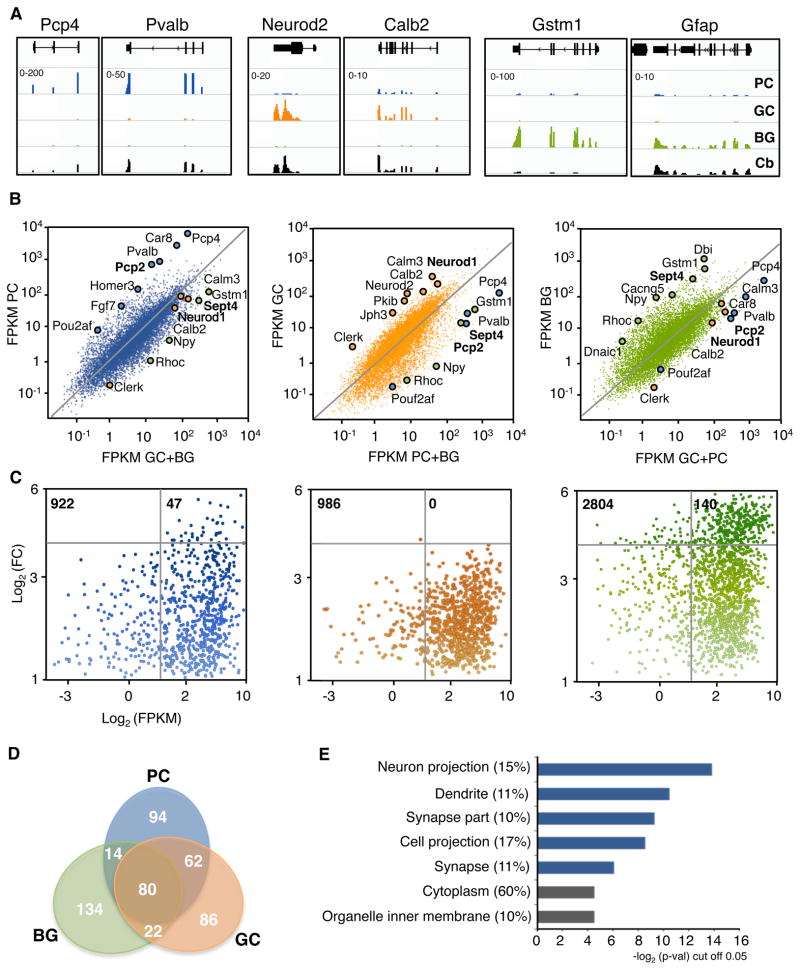
Figure 1. Cell type specific gene expression in cerebellar cell types by TRAP-Seq.
(A) Visualization of FPKM values of TRAP-Seq data of various examples in PC, GC and BG. Pcp4 and Pvalb are enriched in PC (blue), Neurod2 and Calb2 are enriched in GC (orange) and Gstm1 and Gfap are enriched in BG (green). Values of RNA sequencing for these genes from total cerebellum (black) are also shown for comparison. Scale (minimum-maximum) is indicated on the top left corner of PC line for each gene. Windows show the following locations: Pcp4 chr16:96,683,159–96,757,502; Pvalb chr15:78,019,548–78,036,586 ; Neurod2 chr11:98,186,324–98,191,364; Calb2 chr8:112,663,312–112,696,669; Gstm1 chr3:107,814,027–107,821,968; Gfap chr11:102,746,534–102,760,963.
(B) Scatter plots comparing FPKM of TRAP-Seq of an individual cell type [y-axis; blue (left) PC, orange (center) GC, green (right) BG] vs the averaged values of the other two (x-axis). Remarked dots in each panel represent genes enriched in each cell type (PC blue, GC orange and BG green) as previously described by microarray analysis in the literature. Genes in bold indicate the reporter gene of each cell type (n=4 per cell type)
(C) Gene distribution showing enrichment (y-axis; Log2 FC: fold change) and expression values (Log2 FPKM) of enriched genes (> 2 FC) of each cell type compared individually with the other two, and averaged. The number of genes represented are indicated on the top left corner. Intensity of the color represents the level of enrichment. Grey bars delimit genes that are highly enriched (horizontal, cut off: 15 fold) and medium to highly expressed (vertical, cut off: 6 FPKM).
(D) Venn diagram of the 250 most expressed genes of each cell type.
(E) GO analysis of the 94 most expressed genes in PC that are not highly expressed in GC or BG as shown in panel D, p-val cut off 0.05. In parenthesis, percentage of genes that are included in each term. GO terms that explain the main biological features of PCs are highlighted in blue.