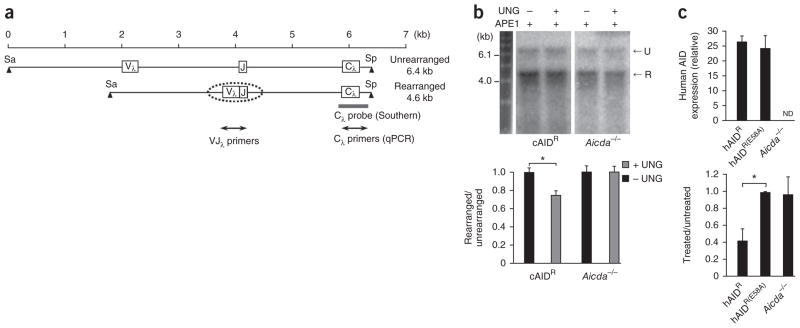
Figure 2.
Uracil residues in the rearranged Igl allele from UNG-deficient DT40 cells. (a) Unrearranged and rearranged alleles. Digestion with the restriction enzymes SacI (Sa) and SpeI (Sp) produces fragments of 6.4 and 4.6 kb. Dotted oval indicates the scope of SHM; gray bar shows the position of the Cλ probe; double-ended arrows indicate the area amplified for quantitative PCR analysis. (b) Southern blot analysis (top) of genomic DNA obtained from cells containing a transgene for overexpression of chicken AID (cAIDR) and from Aicda−/− clones, then treated with APE1 with (+) or without (−) UNG treatment, separated by electrophoresis though an alkaline gel and blotted with the Cλ probe. U, unrearranged; R, rearranged. Below, quantification of band intensity. *P = 0.0001 (two-tailed t-test). Data are representative of six experiments with individual isolation of DNA in each (error bars, s.e.m.). (c) Expression of human AID (top) by DT40 clones transfected with vector encoding wild-type human AID (hAIDR; n = 6 clones) or a catalytically inactive mutant of human AID (hAIDR(E58A); n = 4 clones) or empty vector (Aicda−/−; n = 5 clones), presented relative to the expression of chicken β-actin. ND, not detectable. Below, quantitative PCR analysis of the amplification of VJλ in genomic DNA from treated and untreated samples, presented relative to the amplification of Cλ. *P = 0.0001 (two-tailed t-test). Data are representative of three experiments (average and s.e.m.).