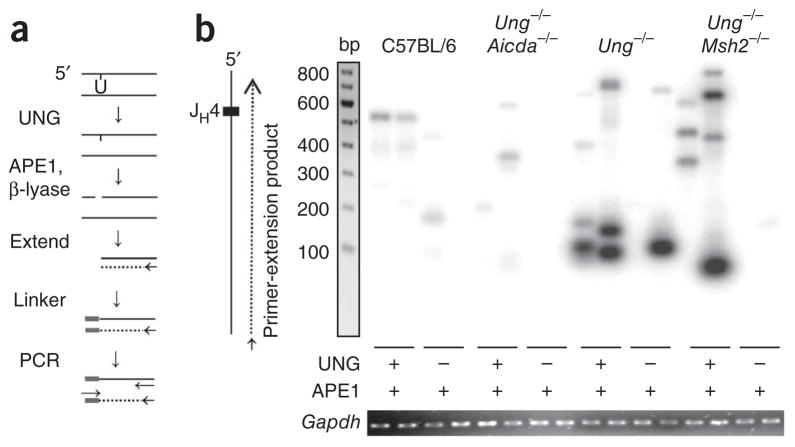
Figure 7.
Detection of uracil residues by ligation-mediated PCR. (a) Experimental design: genomic DNA (100 ng) is treated with UNG, APE1 and DNA polymerase β-lyase to create a blunt 5′ end, then is extended from a primer (arrow) with polymerase to produce a double strand. The product is ligated to a linker and amplified with primers (arrows) for the linker and JH4 intron. (b) Southern blot analysis of DNA obtained from C57BL/6, Ung −/−Aicda−/−, Ung−/− and Ung −/−Msh2−/− B cells, treated as in a (below lanes), amplified by extension initiated ~400 bp downstream of the JH4 gene segment along the nontranscribed strand, separated by gel electrophoresis and hybridized to a JH4 probe. Below, ethidium bromide staining of Gapdh (control for the DNA input used for the initial extension). Data are representative of 50 experiments with amplification of DNA from six to nine mice per strain.