Abstract
Understanding the relationship between life-history variation and population structure in marine invertebrates is not straightforward. This is particularly true of polar species due to the difficulty of obtaining samples and a paucity of genomic resources from which to develop nuclear genetic markers. Such knowledge, however, is essential for understanding how different taxa may respond to climate change in the most rapidly warming regions of the planet. We therefore used over two hundred polymorphic Amplified Fragment Length Polymorphisms (AFLPs) to explore population connectivity at three hierachical spatial scales in the direct developing Antarctic topshell Margarella antarctica. To previously published data from five populations spanning a 1500 km transect along the length of the Western Antarctic Peninsula, we added new AFLP data for four populations separated by up to 6 km within Ryder Bay, Adelaide Island. Overall, we found a nonlinear isolation-by-distance pattern, suggestive of weaker population structure within Ryder Bay than is present over larger spatial scales. Nevertheless, significantly positive F st values were obtained in all but two of ten pairwise population comparisons within the bay following Bonferroni correction for multiple tests. This is in contrast to a previous study of the broadcast spawner Nacella concinna that found no significant genetic differences among several of the same sites. By implication, the topshell's direct-developing lifestyle may constrain its ability to disperse even over relatively small geographic scales.
Introduction
Marine species exhibit extreme heterogeneity in dispersal ability as estimated from genetic data [1] but despite decades of study, the underlying factors are not yet fully understood. One factor that has received a great deal of attention is life-history, since contrasting strategies can either facilitate or hinder dispersal, leading to predictions about the extent to which different species are likely to exhibit population structure [1]–[3]. Specifically, direct developers with restricted dispersal capabilities are hypothesised to be more structured than otherwise ecologically equivalent species with pelagic larvae. In addition, pelagic larval duration is expected to correlate negatively with genetic differentiation.
Although several recent meta-analytical and comparative studies lend support to these theoretical expectations [1]–[9], others have not been able to establish clear links between life-history traits and the strength of population structure [10]–[18]. Such discrepancies could result from any of a large number of potentially confounding factors including larval or adult behaviour [14], [19], the degree of ecological specialisation [11], [20], differences in effective population size [20] or sweepstakes-like reproductive strategies [15]. Moreover, technical factors such as the choice of genetic marker [16], sample size [3] and the completeness of geographic sampling [21] may also play an important role.
In addition to the above factors, our ability to draw general conclusions in respect of the relationship between life history and population structure is also hindered by a bias in the literature towards species from low latitudes [2]. To obtain a more representative view of global patterns therefore requires a broadening of focus to include under-represented geographical and ecological regions such as the poles. Polar species are of particular interest because their development rates are often around 5–10 times slower than equivalent temperate species [22], [23], leading to exceptionally long larval durations in broadcast reproducers. Furthermore the effects of uniquely polar ecological factors such as ice disturbance on population structure have been little studied [24] despite their pervasive and frequent impact on shallow sites [25].
Antarctica provides an unparalleled opportunity to undertake studies of the origins and maintenance of biological diversity, both at the levels of species and populations [26]. Millions of years of isolation from warmer waters to the North by the Antarctic Circumpolar Current have led to the evolution of a diverse and abundant benthic fauna that is both highly endemic and cold adapted [27], [28]. However, parts of Antarctica are now experiencing anthropogenically induced warming at a rate that may soon outstrip the ability of many species to adapt physiologically [29], [30]. For example, sea temperatures have increased by 1°C to the west of the Antarctic Peninsula in the last 50 years [31] and in parallel mean annual air temperatures on the Antarctic Peninsula have increased by as much as 3°C over the same period [32]. This has driven the widespread retreat of glaciers, ice shelf collapse and the exposure of new habitats in both terrestrial and marine locations. In turn, extensive areas of new intense biological productivity have been generated together with associated nearshore benthic ecosystems [33]. This places a premium on genetic studies capable of documenting ‘baseline’ patterns of population structure, which in turn may help to predict the capacity of species or populations to adapt to environmental change [34].
Consistent with predictions based on life history, several Antarctic brooding species have been found to be so highly structured as to invoke cryptic speciation [35]–[39]. However, other brooding species appear unexpectedly homogenous or at least show evidence of gene flow over relatively large geographic scales [40], [41]. Moreover, contradictory findings have also been obtained for several different Antarctic broadcast spawning species [42]–[45]. This suggests the need for comparative studies that are able to control for as many incidental factors as possible, for instance by sampling co-distributed species from the same geographic localities, collecting equal sample sizes of individuals and using the same class of genetic marker.
To evaluate the role of life-history variation within an Antarctic setting, Hoffman et al. [46] recently used Amplified Fragment Length Polymorphisms (AFLPs) to analyse paired populations of the broadcast-spawning Antarctic limpet Nacella concinna and the brooding topshell Margarella antarctica sampled at five locations along the Antarctic Peninsula (see Figure 1 for the sampling sites). The broadcast-spawner was found to be panmictic across most of the peninsula, with only two populations from the extremes of the range, Adelaide and Signy Islands, being genetically differentiated. In contrast, population structure was seven times stronger overall in M.antarctica, with model-based clustering approaches assigning all of the individuals correctly to their source populations based solely on their multilocus genotypes.
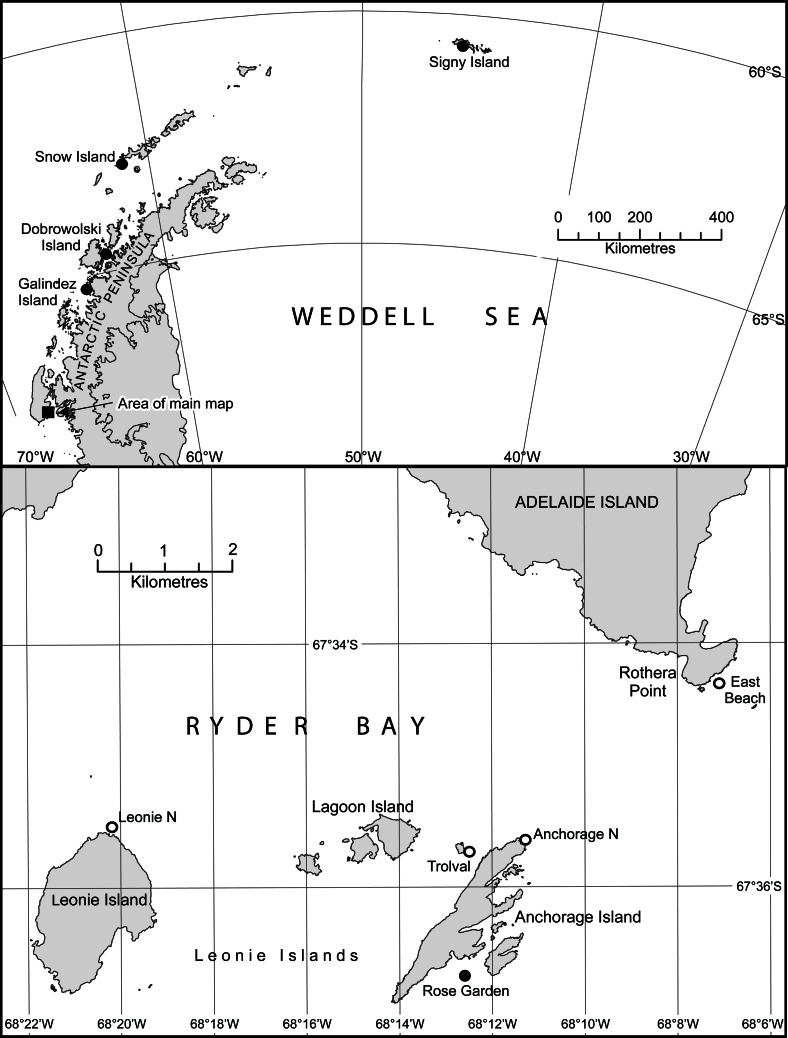
Figure 1. Map showing M. antarctica sampling locations.
Populations sampled and analysed as part of the current study are denoted by white points, while those sampled previously by Hoffman et al. [46] are denoted by black points. The upper panel shows the Antarctic Peninsula and the lower panel shows the Ryder Bay area (Adelaide Island). Figure modified from [46].
An intriguing aspect of the above study was the finding that N.concinna populations from Adelaide Island, at the base of the Antarctic Peninsula, were significantly differentiated from the other Antarctic peninsula populations despite being connected by continuous coastline and this species possessing long-lived planktotrophic veliger larvae [47]. To explore this further, the original AFLP study was extended to include additional N. concinna populations from three localities within Ryder Bay, Adelaide Island (Rothera Point, Leonie and Anchorage Islands), each of which was in turn sub-sampled three times to provide a fine-scale geographic perspective [48]. Surprisingly, limpets sampled from Rothera Point and Leonie Island could not be distinguished on the basis of their AFLP profiles from populations sampled further afield along the Antarctic Peninsula, whereas all the three sub-populations from Anchorage Island showed slight but significant genetic differences. One potential explanation for this is that coastal eddies around Anchorage Island could be advecting larvae back towards the shore, a mechanism invoked to explain local ‘hot spots’ of larval retention encountered in computer simulations [49]. Alternatively, the exposed location of Anchorage Island on the outermost edge of Ryder Bay could predispose it to occasional sporadic larval input from sites to the south [50]. Another possibility relates to the fact that, due to the uneven retreat of coastal glaciers and ice shelves, habitats present at Anchorage Island may be thousands of years older than those around Rothera Point [51]. Consequently, limpet populations across Ryder Bay could have experienced markedly different demographic histories.
The above studies raise additional questions in a comparative context. For example, are M. antarctica populations within Ryder Bay more structured than those of N.concinna as predicted by differences in their life history? Also, does life history influence the pattern of microgeographic population structure, or is this shaped in both species by the same underlying processes? To address these questions, we expanded our previous macrogeographic M. antarctica dataset to incorporate 174 additional individuals sampled from four sites over a 1–6 km spatial scale within Ryder Bay. All 414 individuals were genotyped at 228 polymorphic AFLP loci.
Materials and Methods
Tissue sample collection
M. antarctica samples were collected by SCUBA divers during the austral summer of 1999 from the shallow sublittoral zone off Rothera Point (East Beach), Leonie Island (Leonie North North East) and from Anchorage North and Trolval on Anchorage Island (see Table 1 and Figure 1 for details). Samples were stored in 95% ethanol, initially for four months at −20°C and thereafter at room temperature. Previously published AFLP data from Rose Garden (Anchorage Island), Galindez, Dobrowolski, Snow and Signy Islands [46] were also available for comparison. The combined dataset allowed us to investigate population structure over three spatial scales as follows: (i) Fine-scale (up to 2 km, within Anchorage Island) comprising samples from Rose Garden, Anchorage North and Trolval; (ii) Medium-scale (up to 6 km, among islands within Ryder Bay) comprising samples from Anchorage Island, Rothera Point and Leonie Island; (iii) Large scale (up to 1350 km, along the Antarctic Peninsula) comprising samples from Ryder Bay, Galindez Island, Dobrolowski Island, Snow Island and Signy Island. Geographic distances among these localities are given in Table 2.
Table 1. Details of sampling locations and numbers of M. antarctica individuals collected, including 240 individuals previously sampled by Hoffman et al. [46].
| Region | Site | Population | Latitude (S) | Longitude (W) | No. of samples |
| Ryder Bay | Anchorage Island | Rose Garden | −67.607 | −68.191 | 48 |
| Anchorage North | −67.602 | −68.202 | 44 | ||
| Trolval | −67.608 | −68.218 | 43 | ||
| Rothera Point | East Beach | −67.572 | −68.118 | 43 | |
| Leonie Island | Leonie North | −67.603 | −68.336 | 44 | |
| Galindez Island | – | – | −65.233 | −64.233 | 48 |
| Dobrolowski Island | – | – | −64.917 | −62.607 | 48 |
| Snow Island | – | – | −62.778 | −61.374 | 48 |
| Signy Island | – | – | −60.677 | −45.607 | 48 |
| Total | – | – | – | – | 414 |
The three spatial scales investigated were as follows (i) Fine-scale (within Anchorage Island) comprising samples from Rose Garden, Anchorage North and Trolval; (ii) Medium-scale (among islands within Ryder Bay) comprising samples from Anchorage Island, Rothera Point and Leonie Island; (iii) Large scale (along the Antarctic Peninsula) comprising samples from Ryder Bay, Galindez Island, Dobrolowski Island, Snow Island and Signy Island.
Table 2. Matrix of geographic distances among the nine M. antarctica sampling sites in kilometres.
| Rose Garden | Anchorage North | Trolval | Rothera Point | Leonie Island | Galindez Island | Dobrowolski Island | Snow Island | Signy Island | |
| Rose Garden | 0 | ||||||||
| Anchorage North | 0.66 | 0 | |||||||
| Trolval | 0.71 | 0.64 | 0 | ||||||
| Rothera Point | 2.98 | 2.50 | 3.12 | 0 | |||||
| Leonie Island | 3.62 | 3.56 | 2.99 | 5.70 | 0 | ||||
| Galindez Island | 304.18 | 303.66 | 304.22 | 301.20 | 305.88 | 0 | |||
| Dobrowolski Island | 370.17 | 369.59 | 370.09 | 367.25 | 371.28 | 88.27 | 0 | ||
| Snow Island | 635.07 | 634.53 | 635.07 | 632.10 | 636.57 | 331.84 | 270.97 | 0 | |
| Signy Island | 1342.37 | 1342.10 | 1342.74 | 1339.71 | 1345.37 | 1083.96 | 1076.25 | 866.43 | 0 |
DNA extraction and AFLP genotyping
Total genomic DNA was extracted from a small piece of foot tissue from each individual using a Qiagen DNeasy tissue extraction kit following the manufacturer's recommended protocols. We then used an AFLP protocol adapted from Vos et al. [52] as detailed by Hoffman et al. [53] that employed seven different selective primer combinations (Table 3). PCR products were resolved by electrophoresis through 6% polyacrylamide gels and exposed to X-ray film for five days. These were developed using a universal X-ray developer (Xograph Healthcare Ltd.) and, if required, a second exposure was made for an adjusted time period. All bands in the approximate size range of 75–300 bp were scored manually by an experienced operator (JIH). Only clear bands with minimal size variation were included, these being recorded as 1 = present and 0 = absent. Pairs of bands that were clearly non-independent were scored as single traits. It was assumed that AFLP bands that were the same size across individuals represented homologous markers. In combining our new data with those previously published, we also revisited the original autoradiographs and were able to score a small number of additional loci across all of the samples.
Table 3. Primer combinations used for the AFLP selective amplification and numbers of AFLP bands generated in 414 M. antarctica individuals.
| TaqI primer (5′-3′) | EcoRI primer (5′-3′) | Number of loci | Number of polymorphic loci | % of polymorphic loci |
| GATGAGTCCTGACCGA–CTG | GACTGCGTACCAATTC–AGC | 47 | 42 | 89.4 |
| GATGAGTCCTGACCGA–CGA | GACTGCGTACCAATTC–AGC | 42 | 37 | 86.1 |
| GATGAGTCCTGACCGA–CAG | GACTGCGTACCAATTC–AGC | 30 | 22 | 73.3 |
| GATGAGTCCTGACCGA–CAC | GACTGCGTACCAATTC–AGC | 32 | 25 | 78.1 |
| GATGAGTCCTGACCGA–CCA | GACTGCGTACCAATTC–AAC | 29 | 20 | 69.0 |
| GATGAGTCCTGACCGA–CGA | GACTGCGTACCAATTC–ATG | 44 | 38 | 86.4 |
| GATGAGTCCTGACCGA–CAG | GACTGCGTACCAATTC–ATG | 52 | 44 | 84.6 |
| Total | 276 | 228 | 82.6 |
Data analysis
The program Aflp-Surv [54] was used to calculate global F st together with pairwise F st values among all of the populations. Statistical significance was determined using permutation tests based on 10,000 randomisations of the dataset. Geographic distances among populations were calculated using a Geographic Information System (ESRI ArcGis v 9.2) as described in detail by Hoffman et al. [45]. The significance of correlations between genetic and geographic distance was assessed using Mantel tests with 999 iterations implemented in Genalex v6 [55].
Results
Overall population structure
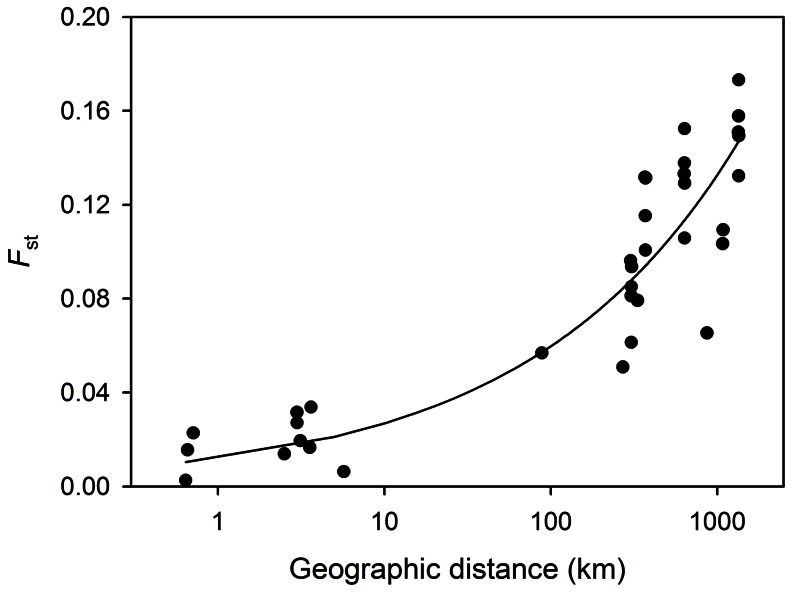
To a previous dataset comprising five Margarella antarctica populations spanning the Antarctic Peninsula [46] we added new AFLP data for four populations within Ryder Bay, Adelaide Island (See Table 1 and Figure 1 for details). The resulting dataset comprised 414 individuals scored for 276 AFLP bands, of which 228 (82.6%) were polymorphic (Table 3). A permutation test for genetic differentiation among all nine populations based on 10, 000 randomisations of the dataset indicated a strong deviation from the null hypothesis of no genetic structure (F st = 0.0872, P<0.0001). Restricting the analysis to the five populations sampled within Ryder Bay, global F st was lower (0.0181) but still highly significant (P<0.0001), indicating the presence of fine-scale population genetic structuring. The relationship between geographic and genetic distance was highly significant overall (Figure 2, Mantel's r = 0.00, n = 9, P<0.001), with a power regression fitting the data considerably better (r 2 = 0.811) than linear, logarithmic or second order polynomial regressions (r 2 = 0.641, 0.777 and 0.782 respectively). Restricting the analysis to populations within Ryder Bay, the isolation-by-distance relationship remained positive but failed to reach statistical significance (Mantel's r = 0.09, n = 5, P = 0.383).
Figure 2. The relationship between geographic and genetic distance (Fst) among nine M. antarctica populations.
To indicate the underlying trend, a power regression was fitted (y = 0.0121×0.3467, r 2 = 0.811), which explains a marginally greater proportion of the variance than logarithmic or second order polynomial regressions (r 2 = 0.777 and 0.782 respectively).
Population structure within Ryder Bay
F st values for each of the pairwise population comparisons within Ryder Bay ranged from 0.003 to 0.034 (Table 4). All but one of these values were individually significant, six out of ten being so at P<0.0001. The only comparison failing to reach significance was that between Anchorage North and Trolval, the two geographically closest populations sampled from a single Island, Anchorage. Following table-wide Bonferroni correction for multiple statistical tests, F st between Leonie Island and Rothera Point also became no longer significant (P = 0.076).
Table 4. Pairwise F st values among M. antarctica sampled from nine different sites (above diagonal).
| Rose Garden | Anchorage North | Trolval | Rothera Point | Leonie Island | Galindez Island | Dobrowolski Island | Snow Island | Signy Island | |
| Rose Garden | * | 0.016 | 0.023 | 0.032 | 0.034 | 0.061 | 0.101 | 0.106 | 0.132 |
| Anchorage North | 0.0002 | * | 0.003 | 0.014 | 0.017 | 0.081 | 0.115 | 0.138 | 0.158 |
| Trolval | <0.0001 | 0.0578 | * | 0.020 | 0.027 | 0.085 | 0.131 | 0.152 | 0.173 |
| Rothera Point | <0.0001 | 0.0005 | <0.0001 | 0.006 | 0.096 | 0.132 | 0.133 | 0.151 | |
| Leonie Island | <0.0001 | <0.0001 | <0.0001 | 0.0022 | * | 0.094 | 0.132 | 0.129 | 0.149 |
| Galindez Island | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | * | 0.057 | 0.079 | 0.109 |
| Dobrowolski Island | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | * | 0.051 | 0.103 |
| Snow Island | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | * | 0.065 |
| Signy Island | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | * |
P-values, calculated using 10,000 permutations of the dataset, are given below the diagonal. All of the F st values were significant at P<0.05, except for that between Anchorage North and Trolval. After table-wide Bonferroni correction for multiple statistical tests, F st the comparison between Rothera Point and Leonie Island also became non-significant.
Discussion
Although studies of the population genetic structure of marine species are commonplace, relatively few have explicitly compared direct with indirect-developers, particularly over multiple geographic scales and using the same methodologies with samples from the same sites. Consequently, we extended previous work on the brooding top shell M. antarctica to allow a comparison over three hierarchical spatial scales with the broadcast-spawning limpet N. concinna. Consistent with expectations, M. antarctica populations were structured throughout Ryder Bay, with the overall magnitude of genetic differentiation being greater than previously found in equivalent comparisons involving N. concinna. This lends further support to the notion that life history could be an important determinant of population structure in many benthic marine species.
Strength and patterns of population genetic structure
Because the previous study of M. antarctica sampled only five populations from the Western Antarctic Peninsula [46], this did not allow inferences to be drawn in respect of geographic scales below 100 km. Specifically, it was unclear whether the isolation-by-distance relationship could be linear, which would imply strong microgeographic population structure, or whether this could instead break down over finer geographic scales. By adding data from four populations within Ryder Bay, we were able to resolve a non-linear profile, with a power regression providing a greatly improved fit to the data relative to a linear one (r 2 = 0.811 versus 0.661, Figure 2). By implication, population structure appears to be far weaker over microgeographic than macrogeographic scales. This is consistent with a recent study of a brooding sea urchin that found genetic differences between patches separated by around ten metres, but little in the way of genetic structure within patches [34]. In contrast, however, distances of several kilometres appear to be just as effective a barrier to dispersal as far larger regions of unsuitable habitat in a direct-developing cushion star [7]. The reasons underlying such interspecific differences remain unclear, partly due to a paucity of empirical data. These may be specific to the species studied or could instead result from generic regional effects such as markedly stronger seasonality and temporally restricted resources, ice scour in polar regions, or the slowed development and delayed maturity characteristic of low temperature species.
Despite population structure being relatively weak within Ryder Bay, global F st was still significantly non-zero (0.0181, P<0.0001) and around twenty times larger than the equivalent value for N. concinna based on the same five populations (0.0009, P = 0.024). Such a marked difference could potentially reflect the influence of Antarctic conditions on the two species' life histories. For example, larval lifetimes tend to be longer in species adapted to cold climates [22], [56], [57], thereby extending the duration of the dispersal phase and hence the scope for gene flow. This leads to the prediction that, if all other factors could be controlled for, differences in population structure between brooders and pelagic developers could be greater at high latitudes than low latitudes. However, set against this, polar marine species tend to have disproportionately long lifespans (e.g. Laternula elliptica can live for up to 36 years [58] and Adamussium colbecki for over 100 years [59]). This could conceivably allow more time for adults of direct-developing gastropod species to migrate from site to site where suitable substratum and depth allow.
A related observation is that M. antarctica and N. concinna not only differed in the strength of population structure, but also in the way in which genetic variation was partitioned within Ryder Bay. All but two pairwise comparisons in M. antarctica were statistically significant following Bonferroni correction for multiple tests, suggesting that population structure although weak may extend across much if not all of the bay. In contrast, most of the N. concinna populations from Ryder Bay were indistinguishable from one another, with only three populations from Anchorage Island revealing significant genetic differences [48]. These contrasting patterns suggest that different factors may be influencing local population structure in the two species. One possibility is that coastal eddies or sporadic larval input from outside the bay could be disproportionately important in the broadcast spawner. To allow generalisation beyond the single comparison we have drawn here, it would be interesting to sample additional brooding and broadcasting species from across the bay, the expectation being that broad similarities should be found within, but not between the two classes of organism.
Another factor that could potentially contribute to population structure in M. antarctica is benthic topology. Although Ryder Bay is only 15–20 km across, it attains a maximum depth of over 500 metres [60], which is beyond the distribution depth of many shallow-water marine species. There is also considerable variation in benthic topology and substratum. The latter could be important because the capacity of M. antarctica to disperse over soft sediments is greatly diminished relative to hard substrata such as bed rock or loose rubble. This hypothesis would be amenable to testing using a ‘landscape genetics’ approach if sufficiently fine-scale data could be collected on depth and substrate distribution across Ryder Bay. It is noteworthy in this context that the only comparison in which we did not obtain a significant F st value prior to Bonferroni correction involved the two geographically closest M. antarctica populations, Trolval and Anchorage North, which were situated less than one kilometre apart on the coast of Anchorage Island. These two sites are separated by rocky coast and around 100 m of soft sediment at depths of 30–40 m (L. Peck pers. obs). However both sites are on the north side of Anchorage Island where steep to vertical rocky slopes descend to beyond 300 m. It is therefore possible that gene transfer between these two sites could take place along the rocky continuum beyond 40 m.
Caveats and future directions
This study was made feasible through the use of AFLPs because these markers are capable of generating large numbers of genome-wide distributed bands in virtually any organism, including diverse Antarctic marine taxa [61], with little need for optimisation. Although we were able to score a large number of bands, however, the dominant nature of these markers leads to increased variance in the estimation of allele frequencies [54]. Furthermore, AFLP bands are typically assumed to be independent whereas in reality mutations, insertions or deletions can generate bands of different size that are linked [62]. This is difficult to control for experimentally, although we attempted to do so by scoring only bands that were clearly independent from one another. Similarly, not all bands of the same size are likely to be homologous [63], [64], although manual scoring may help to alleviate this problem because size homoplasious bands representing different loci may show varying intensities as well as some conformation or sequence dependent differences in electrophoretic mobility [65]. Many of the above factors could be eliminated in future studies by deploying Single Nucleotide Polymorphisms (SNPs), since these markers are codominant and can be mapped to a reference genome to ensure selection of an unlinked, genome-wide distributed panel. Moreover, fully automated genotyping and scoring methods allow SNPs to be genotyped with minimal error [66].
As with any type of genetic marker, AFLPs can also be difficult to compare across species boundaries because different subsets of bands will be generated and these will invariably differ both in number and polymorphic information content. However, we believe this is unlikely to account for the contrasting strength and patterns of genetic structure observed in this study, since the number of informative bands obtained for M. antarctica and N. concinna individuals from Ryder Bay was fairly similar at 189 and 155 respectively, and the smaller panel is still reasonably large. Classical analyses of population structure also assume that all loci are selectively neutral, but some studies have reported conflicting results for different types of marker, suggesting that this may not always be the case [67]. This is implicit in the widespread use of AFLPs for conducting genome scans for ‘outlier loci’ that may be influenced by natural selection [68], [69], although this approach is rapidly being superceded by Restriction Site Associated DNA (RAD) sequencing and allied approaches that draw upon emerging high-throughput sequencing technologies [70]. These could potentially be employed in the future to determine whether outlier loci could be important in the context of this particular study.
Elsewhere, several factors other than life-history have also been invoked to explain the population structure of marine species. For example, Galarza et al. [14] found no relationship between either egg type or larval duration and the strength of genetic structure among seven littoral fish species, leading these authors to suggest that genetic connectivity could be influenced by larval or adult behaviour. Similarly, evidence from two comparative studies [13], [71] suggests that habitat specificity may also have a strong impact upon population structure. Another influential factor could be effective population size [20] since genetic drift occurs more quickly in small populations, suggesting that highly abundant species could be relatively predisposed towards being unstructured. Finally, local extinctions and recolonisations or extreme heterogeneity in the reproductive success of individuals may also lead to temporal fluctuations in population structure [72], [73]. To account for these and other factors presents a major challenge, since this would require both temporal sampling and the inclusion of additional phylogenetically diverse species.
Finally, although our study design allowed us to control for technical factors such as the sampling scheme, choice of genetic marker and the genotyping and scoring protocols, we were not able to include further replicates at the same spatial scale from elsewhere that would help disassociate our findings from any conditions that could be specific to Ryder Bay. Detailed lichenographic studies suggest that the retreat of ice has been uneven across the bay, leaving behind a patchwork of habitats of varying ages [51], so we cannot exclude the possibility that the two species recolonised at different rates or from different sources, or that they could have responded differently to demographic challenges such as bottlenecks induced by the ebb and flow of sea ice. This criticism could be partly addressed by incorporating multi-level sampling from other geographical regions. However, Ryder Bay was an ideal choice for this particular study due to its proximity to the British Antarctic Survey research base at Rothera Point together with detailed knowledge of the local area.
Conclusion
This study extended previous work on the direct developing top shell M. antarctica to include a microgeographic component. Overall, a strong but nonlinear isolation-by-distance pattern was detected, indicating relatively weak population structure over scales of 1–6 km. Nevertheless, this was consistent across the bay and greater in magnitude than previously documented for the broadcast spawner N. concinna. By implication, life history variation, specifically protected versus broadcast reproductive modes, may impact the population structure of benthic marine species over multiple geographic scales. This could have implications for understanding how Antarctic marine invertebrates may respond to climate change, since dispersal is a key means by which locally extirpated populations might be replenished from adjacent locations.
Acknowledgments
Samples were collected during the British Antarctic Survey (BAS) Peninsula Geneflow cruise in 1999. We thank the dive team, officers and crew of RRS Bransfield for their support in the collection of these samples and the late Martin White for considerable help and support with sample design and logistical aspects. We are also grateful to Peter Fretwell for providing a matrix of geographic distances among populations and for generating the map of sampling locations. We finally thank two anonymous reviewers for comments that improved the final manuscript.
Funding Statement
JIH was supported by a Natural Environment Research Council (NERC) British Antarctic Survey (BAS) Strategic Alliance Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kinlan BP, Gaines SD (2003) Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84: 2007–2020. [Google Scholar]
- 2. Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE (2008) Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proceedings of the Royal Society of London Series B-Biological Sciences 275: 1803–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selkoe KA, Toonen RJ (2011) Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Marine Ecology Progress Series 436: 291–305. [Google Scholar]
- 4. Arndt A, Smith MJ (1998) Genetic diversity and population structure in two species of sea cucumber: differing patterns according to mode of development. Molecular Ecology 7: 1053–1064. [Google Scholar]
- 5. Doherty PJ, Planes S, Mather P (2005) Gene flow and larval duration in seven species of fish from the great barrier reef. Ecology 76: 2373–2391. [Google Scholar]
- 6. Watts PC, Thorpe JP (2006) Influence of contrasting larval development types on the population-genetic structure of cheilostome bryozoans. Marine Biology 149: 1093–1101. [Google Scholar]
- 7. Sherman CDH, Hunt A, Ayre DJ (2008) Is life history a barrier to dispersal? Contrasting patterns of genetic differentiation along an oceanographically complex coast. Biological Journal of the Linnean Society 95: 106–111. [Google Scholar]
- 8. Underwood JN, Smith LD, Van Oppen MJH, Gilmour JP (2009) Ecologically relevant dispersal of corals on isolated reefs: implications for managing resilience. Ecological Applications 19: 18–29. [DOI] [PubMed] [Google Scholar]
- 9. Nishikawa A, Katoh M, Sakai K (2003) Larval settlement rates and gene flow of broadcast-spawning (Acropora tenuis) and planula brooding (Stylophora pistillata) corals. Marine Ecology Progress Series 256: 87–97. [Google Scholar]
- 10. Ayre DJ, Hughes TP (2000) Genotypic diversity and gene flow in brooding and spawning corals along the Great Barrier Reef, Australia. Evolution 5: 1590–1605. [DOI] [PubMed] [Google Scholar]
- 11. Bay LK, Crozier RH, Caley MJ (2006) The relationship between population genetic structure and pelagic larval duration in coral reef fishes on the Great Barrier Reef. Marine Biology 149: 1247–1256. [Google Scholar]
- 12. Richards VP, Thomas JD, Stanhope MJ, Shivji MS (2007) Genetic connectivity in the Florida reef system: comparative phylogeography of commensal invertebrates with contrasting reproductive strategies. Molecular Ecology 16: 139–157. [DOI] [PubMed] [Google Scholar]
- 13. Ayre DJ, Minchinton TE, Perrin C (2009) Does life history predict past and current connectivity for rocky intertidal invertebrates across a marine biogeographic barrier? Molecular Ecology 18: 1887–1903. [DOI] [PubMed] [Google Scholar]
- 14. Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, et al. (2009) The influence of oceanographic fronts and early life-history traits on connectivity among littoral fish species. Proceedings of the National Academy of Sciences of the United States of America 106: 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee HJ, Boulding EG (2009) Spatial and temporal population genetic structure of four northeastern Pacific littorinid gastropods: the effect of mode of larval development on variation at one mitochondrial and two nuclear DNA markers. Molecular Ecology 18: 2165–2184. [DOI] [PubMed] [Google Scholar]
- 16. Weersing K, Toonen RJ (2009) Population genetics, larval dispersal, and connectivity in marine systems. Marine Ecology Progress Series 393: 1–12. [Google Scholar]
- 17. Kelly RP, Palumbi SR (2010) Genetic Structure Among 50 Species of the Northeastern Pacific Rocky Intertidal Community. PLoS One 5: e8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riginos C, Douglas KE, Jin Y, Shanahan DF, Treml EA (2011) Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography 34: 566–575. [Google Scholar]
- 19. Temby N, Miller K, Mundy C (2007) Evidence of genetic subdivision among populations of blacklip abalone (Haliotis rubra Leach) in Tasmania. Marine and Freshwater Research 58: 733–742. [Google Scholar]
- 20. Faurby S, Barber PH (2012) Theoretical limits to the correlation between pelagic larval duration and population genetic structure. Molecular Ecology 21: 3419–3432. [DOI] [PubMed] [Google Scholar]
- 21. Jenkins DG, Carey M, Czerniewska J, Fletcher J, Hether T, et al. (2010) A meta-analysis of isolation by distance: relic or reference standard for lanscape genetics? Ecography 33: 315–320. [Google Scholar]
- 22. Peck LS, Clarke A, Chapman AL (2006) Metabolism and development of pelagic larvae of Antarctic gastropods with mixed reproductive strategies. Marine Ecology Progress Series 318: 213–220. [Google Scholar]
- 23. Peck LS, Powell DK, Tyler PA (2007) Very slow development in two Antarctic bivalve molluscs, the infaunal clam, Laternula elliptica and the scallop Adamussium colbecki . Marine Biology 150: 1191–1197. [Google Scholar]
- 24. Harper L, Clark MS, Hoffman JI, Phillip EER, Peck L, et al. (2012) Iceberg scour and shell damage in the Antarctic bivalve Laternula elliptica . PLoS ONE 7: e46341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown KM, Fraser KPP, Barnes DKA, Peck LS (2004) Ice scour frequency dictates Antarctic shallow-water community structure. Oecologia 141: 121–129. [DOI] [PubMed] [Google Scholar]
- 26. Clarke A (2000) Evolution in the cold. Antarctic Science 112: 257. [Google Scholar]
- 27. Clarke A, Johnston N (2003) Antarctic marine benthic diversity. Oceanography and Marine Biology: Annual Review 41: 47–114. [Google Scholar]
- 28. Fraser CI, Nikula R, Ruzzante DE, Waters JM (2012) Poleward bound: biological impacts of Southern Hemisphere glaciation. Trends in Ecology and Evolution 27: 462–470. [DOI] [PubMed] [Google Scholar]
- 29. Peck LS (2005) Prospects for survival in the Southern Ocean; vulnerability of benthic species to temperature change. Antarctic Science 17: 495–505. [Google Scholar]
- 30. Peck LS (2011) Organisms and responses to environmental change. Marine Genomics 4: 237–304. [DOI] [PubMed] [Google Scholar]
- 31. Meredith MP, King JC (2005) Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophysical Research Letters 32: L19604. [Google Scholar]
- 32. King JC, Turner J, Marshall GJ, Connolley WM, Lachlan-Cope TA (2003) Antarctic Peninsula climate variability and its causes as revealed by instrumental records. Antarctic Research Series 79: 17–30. [Google Scholar]
- 33. Peck LS, Barnes DKA, Cook AJ, Fleming AH, Clarke A (2010) Negative feedback in the cold: ice retreat produces new carbon sinks in Antarctica. Global Change Biology 16: 2614–2623. [Google Scholar]
- 34. Ledoux J-B, Tarnowska K, Gerard K, Lhuillier E, Jacquemin B, et al. (2012) Fine-scale spatial genetic structure in the brooding sea urchin Abatus cordatus suggests vulnerability of the Southern Ocean marine invertebrates facing global change. Polar Biology 35: 611–623. [Google Scholar]
- 35. Wilson NG, Hunter RL, Lockhart SJ, Halaynch KM (2007) Multiple lineages and absence of panmixia in the ‘cirumpolar’ crinoid Promachocrinus kerguelensis from the Atlantic sector of Antarctica. Marine Biology 152: 895–904. [Google Scholar]
- 36. Mahon AR, Arango CP, Halynch KM (2008) Genetic diversity of Nymphon (Arthropoda: Pycnogonida: Nymphonidae) along the Antarctic Peninsula with a focus on Nymphon australe Hodgson 1902. Marine Biology 155: 315–323. [Google Scholar]
- 37. Linse K, Cope T, Lorz A-N, Sands C (2009) Is the Scotia Sea a centre of Antarctic marine diversification? Some evidence of cryptic speciation in the circum-Antarctic bivalve Lissarca notorcadensis (Arcoidea: Philobryidae). Polar Biology 30: 1059–1068. [Google Scholar]
- 38. Thornhill DJ, Mahon AR, Norenburg JL, Halanych KM (2008) Open-ocean barriers to dispersal: a test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae). Molecular Ecology 17: 5104–5117. [DOI] [PubMed] [Google Scholar]
- 39. Krabbe K, Leese F, Mayer C, Tollrian R, Held C (2010) Cryptic mitochondrial lineages in the widespread pycnogonid Colossendeis megalonyx Hoek, 1881, from Antarctic and Subantarctic waters. Polar Biology 33: 281–292. [Google Scholar]
- 40. Hunter RL, Halynch KM (2008) Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the Drake Passage in the Southern Ocean. Journal of Heredity 99: 137–148. [DOI] [PubMed] [Google Scholar]
- 41. Leese F, Agrawal S, Held C (2010) Long-distance island hopping without dispersal stages: transportation across major zoogeographic barriers in a Southern Ocean isopod. Naturwissenschaften 97: 583–594. [DOI] [PubMed] [Google Scholar]
- 42. Rogers AD, Clarke A, Peck LS (1998) Population genetics of the Antarctic heteronemertean Parbolasia corrugatus from the South Orkney Islands. Marine Biology 131: 1–13. [Google Scholar]
- 43. Matschiner M, Hanel R, Salzburger W (2009) Gene flow by larval dispersal in the Antarctic notothenioid fish Gobionotothen gibberifrons . Molecular Ecology 18: 2574–2587. [DOI] [PubMed] [Google Scholar]
- 44. Hunter RL, Halynch KM (2010) Phylogeography of the Antarctic planktotrophic brittle star Ophionotus victoriae reveals genetic structure inconsistent with early life history. Marine Biology 157: 1693–1704. [Google Scholar]
- 45. Hoffman JI, Peck LS, Linse K, Clarke A (2011) Strong population structure in a broadcast-spawning Antarctic marine invertebrate. Journal of Heredity 102: 55–66. [DOI] [PubMed] [Google Scholar]
- 46. Hoffman JI, Clarke A, Linse K, Peck LS (2011) Effects of brooding and broadcasting reproductive modes on the population structure of two Antarctic gastropod molluscs. Marine Biology 158: 287–296. [Google Scholar]
- 47. Bowden DA, Clarke A, Peck LS, Barnes DKA (2006) Antarctic sessile marine benthos: colonisation and growth on artificial substrata over three years. Marine Ecology Progress Series 316: 1–16. [Google Scholar]
- 48. Hoffman JI, Clarke A, Clark MS, Fretwell P, Linse K (2012) Unexpected fine-scale population structure in a broadcast-spawning Antarctic marine mollusc. PLoS ONE 7: e32415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siegel DA, Mitarai S, Costello CJ, Gaines SD, Kendall BE, et al. (2008) The stochastic nature of larval connectivity among nearshore marine populations. Proceedings of the National Academy of Sciences of the United States of America 105: 8974–8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Savidge DK, Amft JA (2009) Circulation on the West Antarctic Peninsula derived from 6 years of shipboard ADCP transects. Deep-Sea Research I 56: 1633–1655. [Google Scholar]
- 51. Golledge NR, Everest JD, Bradwell T, Johnson JS (2010) Lichenometry on Adelaide Island, Antarctic Peninsula: size-frequency studies, growth rates and snowpatches. Geografiska Annaler: Series A, Physical Geography Special Issue: Lichenometry in Subpolar Environments 92: 111–124. [Google Scholar]
- 52. Vos P, Hogers R, Bleker M, Reijans M, Van de Lee T, et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoffman JI, Peck LS, Hillyard G, Zieritz A, Clark MS (2010) No evidence for genetic differentiation between Antarctic limpet Nacella concinna morphotypes. Marine Biology 157: 765–778. [Google Scholar]
- 54.Vekemans X (2002) AFLP-SURV version 1.0. Distributed by the author. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Belgium. [Google Scholar]
- 55. Peakall R, Smouse PE (2005) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Moleular Ecology Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bosch I, Beauchamp KA, Steele ME, Pearse JS (1987) Development, metamorphosis, and seasonal abundance of embryos and larvae of the Antarctic sea urchin Sterechinus neumayeri . Biological Bulletin 173: 126–135. [DOI] [PubMed] [Google Scholar]
- 57. Hoegh-Guldberg O, Pearse JS (1995) Temperature, food availability, and the development of marine invertebrate larvae. American Zoologist 35: 415–425. [Google Scholar]
- 58. Philipp E, Brey T, Pörtner H-O, Abele D (2004) Chronological and physiological ageing in a polar and a temperate mud clam. Mechanisms of ageing and Development 126: 598–609. [DOI] [PubMed] [Google Scholar]
- 59. Berkman PA, Cattaneo-Vietti R, Chiantore M, Howard-Williams C (2004) Polar emergence and the influence of increased sea-ice extent on the Cenozoic biogeography of pectinid molluscs in Antarctic coastal areas. Deep-Sea Research II 51: 1839–1855. [Google Scholar]
- 60. Clarke A, Mereditha MP, Wallacea MI, Brandon MA, Thomas DN (2008) Seasonal and interannual variability in temperature, chlorophyll and macronutrients in northern Marguerite Bay, Antarctica. Deep Sea Research Part II: Topical Studies in Oceanography 55: 1988–2006. [Google Scholar]
- 61. Hoffman J, Clark MS, Amos W, Peck LS (2012) Widespread amplification of Amplified Fragment Length Polymorphisms (AFLPs) in marine Antarctic animals. Polar Biology 35: 919–929. [Google Scholar]
- 62. Simmons MP, Zhang LB, Webb CT, Muller K (2007) A penalty of using anonymous dominant markers (AFLPs, ISSRs, and RAPDs) for phylogenetic inference. Molecular Phylogenetics and Evolution 42: 528–542. [DOI] [PubMed] [Google Scholar]
- 63. O'Hanlon PC, Peakall R (2000) A simple method for the detection of size homoplasy among amplified fragment length polymorphism fragments. Molecular Ecology 9: 815–816. [DOI] [PubMed] [Google Scholar]
- 64. Althoff DM, Gitzendanner MA, Segraves KA (2007) The utility of amplified fragment length polymorphisms in phylogenetics: a comparison of homology within and between genomes. Systematic Biology 56: 477–484. [DOI] [PubMed] [Google Scholar]
- 65. Dasmahapatra KK, Hoffman JI, Amos W (2009) Pinniped phylogenetic relationships inferred using AFLP markers. Heredity 103: 168–177. [DOI] [PubMed] [Google Scholar]
- 66. Hoffman JI, Tucker R, Clark MS, Forcada J, Slate J (2012) Rates of assay success and genotyping error when single nucleotide polymorphism genotyping in non-model organisms: a case study in the Antarctic fur seal. Molecular Ecology Resources 12: 861–872. [DOI] [PubMed] [Google Scholar]
- 67. Hellberg ME, Burton RS, Neigel JE, Palumbi SR (2002) Genetic assessment of connectivity among marine populations. Bulletin of Marine Science 70: 273–290. [Google Scholar]
- 68. Bensch S, Akesson M (2005) Ten years of AFLP in ecology and evolution: why so few animals? Molecular Ecology 14: 2899–2914. [DOI] [PubMed] [Google Scholar]
- 69. Bonin A, Ehrich D, Manel S (2007) Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Molecular Ecology 16: 3737–3758. [DOI] [PubMed] [Google Scholar]
- 70. Davey JW, Blaxter ML (2011) RADSeq: next-generation population genetics. Briefings in Functional Genomics 9: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bird CE, Holland BS, Bowen BW, Toonen RJ (2007) Contrasting phylogeography in three endemic Hawaiian limpets (Cellana spp.) with similar life histories. Molecular Ecology 16: 3173–3186. [DOI] [PubMed] [Google Scholar]
- 72. Hedgecock D (1994) Temporal and spatial genetic structure of marine animal populations in the California current. CalCOFI Rep 35: 73–81. [Google Scholar]
- 73. Piertney SB, Carvalho GR (1995) Microgeographic genetic differentiation in the intertidal isopod Jaera albifrons Leach. II. Temporal variation in allele frequencies. Journal of Experimental Marine Biology and Ecology 188: 277–288. [Google Scholar]