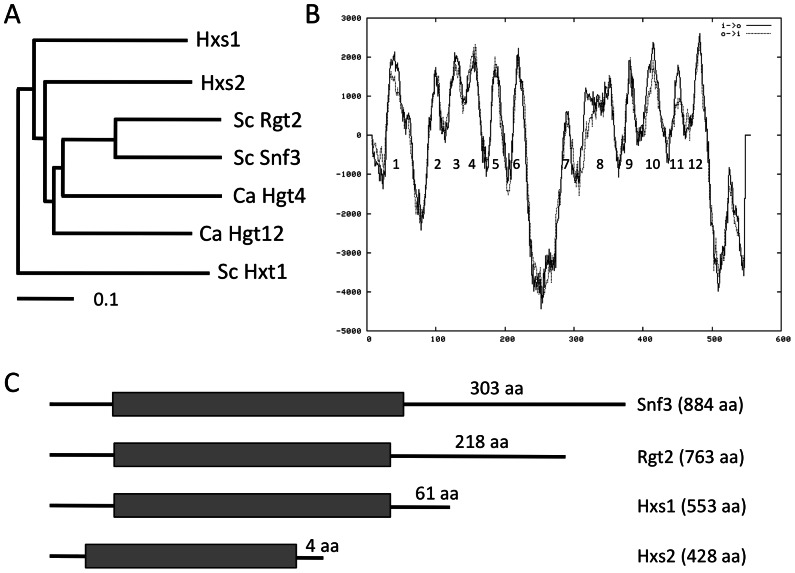
Figure 1. Identification of Snf3/Rgt2 homologs in Cryptococcus.
A. Phylogenetic tree of proteins from C. albicans (Hgt4 and Hgt12) and C. neoformans (Hxs1 and Hxs2) that share high homology with glucose sensors Snf3 and Rgt2 in S. cerevisiae. The full-length protein sequences were used for alignment using ClustalX. The hexose transporter Hxt1 in S. cerevisiae was used as an out-group. B. Predicted topology of the deduced Hxs1 amino acid sequence based on the TMpred program. Hydropathy values are on the y-axis, and the residue numbers are on the x-axis. The predicted transmembrane domains (TM1 to 12) are numbered. C. Schematic models of the primary protein structures of Snf3, Rgt2, Hxs1 and Hxs2. The black boxes represent transmembrane regions. The total number of amino acids for each protein and its C-terminal tail are indicated.