Table 3.
Summary of key metabolic genes identified in sequence assemblies of the predominant archaeal populations present across sites, which exhibited a wide range in pH, dissolved sulfide, and dissolved oxygen (see Table 1).
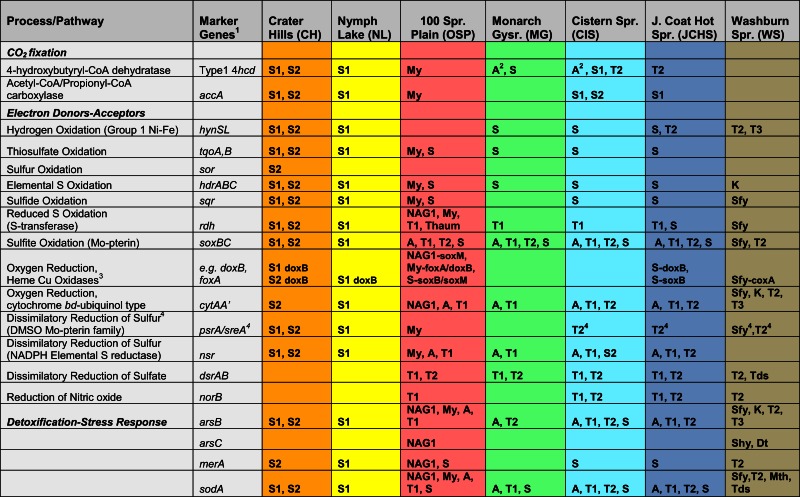 |
Population types: S1 and S2, Sulfolobales Types 1 and 2, respectively; My, M. yellowstonensis; A, Desulfurococcales (Acidilobus sp.); T1, T2, T3, Thermoproteales. Types 1, 2, and 3, respectively; NAG1, “Novel Archaea Group I” (Candidate phylum Geoarchaeota; Kozubal et al., 2013); K, Korarchaeota; Sfy, Sulfurihydrogenibium-like; Dt, Dictyoglomus-like; Mth, Methanosarcinales; Tds, Thermodesulfobacteria.
1“High confidence” sequence matches to marker genes that code for proteins with high specificity for possible pathway. Query sequences used to search for specific functional genes given in Table S3 in Supplementary Material (Inskeep et al., 2013). Genes not present in these sites = amoA, nifH, mcrA, nirK, nirS, nosZ.
2Specific heme Cu oxidases (subunit 1) are listed for each population type.
3Uncharacterized DMSO-Mo-pterins belonging to the Thermoproteales (T2) and Sulfurihydrogenibium (Sfy) populations are listed here.
