Abstract
The disease take-all, caused by the fungus Gaeumannomyces graminis, is one of the most destructive root diseases of wheat worldwide. Breeding resistant cultivars is an effective way to protect wheat from take-all. However, little progress has been made in improving the disease resistance level in commercial wheat cultivars. MYB transcription factors play important roles in plant responses to environmental stresses. In this study, an R2R3-MYB gene in Thinopyrum intermedium, TiMYB2R-1, was cloned and characterized. The gene sequence includes two exons and an intron. The expression of TiMYB2R-1 was significantly induced following G. graminis infection. An in vitro DNA binding assay proved that TiMYB2R-1 protein could bind to the MYB-binding site cis-element ACI. Subcellular localization assays revealed that TiMYB2R-1 was localized in the nucleus. TiMYB2R-1 transgenic wheat plants were generated, characterized molecularly, and evaluated for take-all resistance. PCR and Southern blot analyses confirmed that TiMYB2R-1 was integrated into the genomes of three independent transgenic wheat lines by distinct patterns and the transgene was heritable. Reverse transcription–PCR and western blot analyses revealed that TiMYB2R-1 was highly expressed in the transgenic wheat lines. Based on disease response assessments for three successive generations, the significantly enhanced resistance to take-all was observed in the three TiMYB2R-1-overexpressing transgenic wheat lines. Furthermore, the transcript levels of at least six wheat defence-related genes were significantly elevated in the TiMYB2R-1 transgenic wheat lines. These results suggest that engineering and overexpression of TiMYB2R-1 may be used for improving take-all resistance of wheat and other cereal crops.
Key words: Gaeumannomyces graminis var. tritici, MYB transcription factor, take-all resistance, Thinopyrum intermedium, transformation, Triticum aestivum.
Introduction
Wheat (Triticum aestivum) is one of the most important food crops in the world. Root diseases have considerable economic impacts on wheat production. The disease known as take-all, caused by the necrotrophic fungus Gaeumannomyces graminis var. tritici (Ggt), is one of the most destructive root diseases of wheat worldwide (Gutteridge et al., 2003; Daval et al., 2011). The disease begins by Ggt hyphae penetrating the cortical cells of the root and progresses upwards into the base of the stem, and even leads to premature death of the infected plant. The symptoms of the disease are manifested as black lesions on the roots. Symptoms on above-ground parts of the infected plant include stunting, premature ripening, and white heads (bleached white and empty spikes) (Cook, 2003; Guilleroux and Osbourn, 2004). Take-all can affect the quality and yield of wheat [i.e. yield losses of 40–60% (Gutteridge et al., 2003)]. Take-all also impacts the production of barley (Hordeum vulgare) and triticale (Secale×Triticum) (Gutteridge et al., 2003; Bithell et al., 2011).
Breeding wheat varieties with resistance is the most promising and reliable way to protect wheat from take-all. Since no effective resistance has been identified in wheat accessions (Gutteridge et al., 2003; Yang et al., 2011), to breed take-all-resistant wheat varieties using traditional methods does not appear to be feasible. The advent of genetic engineering and its application to the production of crops make it possible to generate wheat materials with resistance to this disease.
Plants have evolved sophisticated defence mechanisms to cope with pathogens. Many transcription factor (TF) families have been shown to play important roles in defence responses through regulating the expression of defence-related genes. MYB TFs are characterized by a MYB domain conferring an ability to bind the cis-acting elements of targeted genes (Dubos et al., 2010). Based on the numbers of adjacent repeats in the MYB domain, MYB proteins are classified into four subfamilies, namely R1MYB, R2R3-MYB, 3R-MYB, and 4R-MYB (Dubos et al., 2010). Since the first plant MYB gene required for synthesis of anthocyanins, C1, was isolated from maize (Zea mays) (Paz-Ares et al., 1987), a large number of MYB proteins have been identified in various plant species, namely Arabidopsis thaliana, rice (Oryza sativa), maize (Zea mays), cotton (Gossypium hirsutum), grapevine (Vitis vinifera L.), poplar (Populus tremuloides), apple (Malus domestica), wheat, and Avicennia marina (Rabinowicz et al., 1999; Cedroni et al., 2003; Dias et al., 2003; Chen et al., 2006; Dubos et al., 2010; Zhang et al., 2011; Ganesan et al., 2012). MYB proteins perform diverse biological functions in the cell cycle and in development, regulation of primary and secondary metabolism, and abiotic stress response (Ma et al., 2009; Seo et al., 2009; Dubos et al., 2010; Seo and Park, 2010; He et al., 2011; Xue et al., 2011; Zhang et al., 2011, 2012; Qin et al., 2012; Romano et al., 2012; Shen et al., 2012). Some plant MYB proteins are involved in defence responses to pathogens. For instance, Arabidopsis R2R3-MYB proteins, including AtMYB108 and AtMYB96, participate in disease resistance (Mengiste et al., 2003; Seo and Park, 2010). Our previous study showed that overexpression of a wheat MYB gene TaPIMP1 enhanced resistance to biotic and abiotic stresses in transgenic tobacco and wheat (Liu et al., 2011; Zhang et al., 2012).
Thinopyrum intermedium (Agropyron intermedium, intermedium wheatgrass; 2n=42), a wild relative of wheat, is naturally resistant to wheat diseases, such as leaf rust, yellow rust, and stem rust (Cauderon et al., 1973), Wheat streak mosaic virus (WSMV; Sharma et al., 1984), Barley yellow dwarf virus (BYDV; Sharma et al., 1984), Fusarium head blight (FHB; Fedak and Han, 2005), and eyespot (Li et al., 2005). The resistance to wheat rusts, BYDV, WSMV, FHB, and eyespot has been introgressed into the wheat background and characterized by different groups (Cauderon et al., 1973; Xin et al., 1991; Banks et al., 1995; Sharma et al., 1995; Fedak and Han, 2005; Li et al., 2005). However, it is not clear whether R2R3 MYB TFs in T. intermedium are involved in defence responses. Therefore, a study of the species-specific MYB genes may provide insights into T. intermedium defence mechanisms.
In this study, the first R2R3-MYB gene isolated from T. intermedium, TiMYB2R-1, was cloned. Its R2R3-MYB activity was confirmed by subcellular localization and cis-element binding activity assays. The functional characteristics of TiMYB2R-1 in defense responses to take-all pathogen Ggt were also explored through its expression in generated transgenic wheat lines. The results showed that the ectopic expression of TiMYB2R-1 significantly increased resistance to take-all in transgenic wheat.
Materials and methods
Plant and fungal materials and treatments
Thinopyrum intermedium cultivar (cv.) Z1146 was provided by Dr Lihui Li, Institute of Crop Science, CAAS. The wheat cv. Yangmai 12, provided by Lixiahe Agricultural Institute of Jiangsu, China, was used as the recipient of TiMYB2R-1 transformation. Yangmai 12 is a Chinese commercial wheat variety with susceptibilty to Ggt and is a good material for this study.
The fungal pathogen Ggt XNQS-2 was isolated, identified, and provided by Dr Yang Wang, College of Plant Protection, Northwest A&F University, China.
For inoculation, the Ggt fungus was cultured on potato dextrose agar (PDA) at 25 °C for ~10 d, then 1cm2 plugs from the edge of Ggt colonies were placed onto the surface of sand in pots. One seed germinated for 2 d was put on the top of each Ggt plug, and covered with 2cm of sand. The plants were cultured in a growth chamber at a 23 °C, 14h light/15 °C, 10h dark regime at 70% relative humidity. The roots were collected at 0, 4, 7, 14, and 21 days post-inoculation (dpi) for RNA extraction.
DNA and RNA extraction and first-strand cDNA synthesis
Genomic DNA was extracted from leaf tissues of T. intermedium Z1146 or wheat as described by Sharp et al. (1988). Total RNA was extracted from roots of T. intermedium or wheat using TRIZOL reagent (Invitrogen), and then subjected to RNase-free DNase I (TaKaRa) treatment and purification.
A 5 µg aliquot of RNA per sample was used to synthesize the first-strand cDNA using a Superscript II First-Strand Synthesis Kit for RT-PCR (Invitrogen).
Cloning and sequence analysis of the TiMYB2R-1 gene
Based on the sequence of the wheat MYB gene TaPIMP1 (accession no. EF587267), a pair of primers (MYB-OF, 5’-ACTCGC GTACGTCTTCCTGA-3’; and MYB-OR, 5’-GCGCTCTAGTTA AGTTCATCGTC-3’) was designed and used to amplify the full-length cDNA sequence of the MYB gene TiMYB2R-1 from cDNA of T. intermedium Z1146 roots at 4 d post-challenge with Ggt. The PCR fragment corresponding to TiMYB2R-1 was excised, cloned, and its sequence was analysed. The cDNA sequence of TiMYB2R-1 of 1038bp in length was deposited in the National Center for Biotechnology Information (NCBI) with accession number JX683795. TiMYB2R-1 contains an open reading frame (ORF) of 972bp (NCBI accession no. JQ663861). The genomic sequence of TiMYB2R-1 was amplified from genomic DNA of Z1146 using the primers MYB-OF and MYB-OR, then cloned and sequenced. The genomic sequence was deposited in the NCBI with accession no. JX683794.
DNA and protein sequences were analysed using DNAMAN software, DNASTAR software, and BLAST online (http://www.ncbi.nlm.gov/blast).
Subcellular localization of TiMYB2R-1
The coding region of TiMYB2R-1 without the stop codon was amplified using gene-specific primers with HindIII and XbaI restriction sites (TIM-NLF, 5’-GaagcttATGGACATGGACAAGGAGTA-3’; and TIM-NLR, 5’-CtctagaTCAGCAGTAAATGTCCTCTAG-3’). TiMYB2R-1 was fused in-frame to the 5’ terminus of the green fluorescent protein (GFP) gene in the ph16318 vector (Dr Daowen Wang, CAS), and controlled by the Cauliflower mosaic virus (CaMV) 35S promoter. The resulting TiMYB2R-1-GFP fusion construct or GFP alone were transformed separately into white onion epidermal cells using a PDS-1000/He gene gun (Bio-Rad, USA) at 1100 psi. After incubation for 40h at 25 °C, GFP fluorescence in the transformed onion cells was observed under 488nm excitation using a confocal laser scaning microscope (Zeiss LSM 700, Germany) with a Fluar 10×/0.50 M27 objective lens and a SP640 filter.
The cis-element binding assay of TiMYB2R-1
Previously R2R3-MYB proteins were shown to bind to MYB-binding site (MBS) AC cis-elements including ACI (core sequence: ACCTACC, Patzlaff et al., 2003). To investigate if TiMYB2R-1 protein binds to the MBS ACI cis-element, the ORF sequence of TiMYB2R-1 was subcloned in-frame to the 3’ terminus of a glutathione S-transferase (GST) gene in the pGEX-4T-1 vector (GE Amersham), resulting in the recombinant expression vector pGST-TiMYB2R-1. The pGST-TiMYB2R-1 construct was transformed into competent cells of Escherichia coli BL21. The recombinant protein GST–TiMYB2R-1 was expressed after induction with 0.3mM isopropyl-β-d-thiogalactopyranoside for 6h at 16°C, and purified using a MicroSpin module (GE Amersham).
The forward and reverse oligonucleotides containing the MBS ACI cis-element (Zhang et al., 2012) were synthesized and used to prepare the probe. Electrophoretic mobility shift assays (EMSAs) were conducted following the protocol of Zhang et al. (2012). A 1 µg aliquot of the ACI probe plus 3 µg of purified GST–TiMYB2R-1 or GST protein alone were mixed with the binding buffer.
TiMYB2R-1 transformation vector and wheat transformation
The full ORF sequence of the TiMYB2R-1 gene was subcloned into the SmaI and SacI sites of a plant expression vector pAHC25 (Christensen and Quail, 1996), resulting in the transformation vector pA25-TiMYB2R-1. It contained a Ubi::TiMYB2R-1-Tnos chimera, where TiMYB2R-1 was driven by the maize ubiquitin (Ubi) promoter and terminated by the terminator of the Agrobacterium tumefaciens nopaline synthase gene (Tnos). In all, 1200 immature embryos of wheat Yangmai 12 were transformed by biolistic bombardment using pA25-TiMYB2R-1. All aspects of transformation were conducted according to the protocol described by Chen et al. (2008).
PCR detection of TiMYB2R-1 transgenic wheat plants
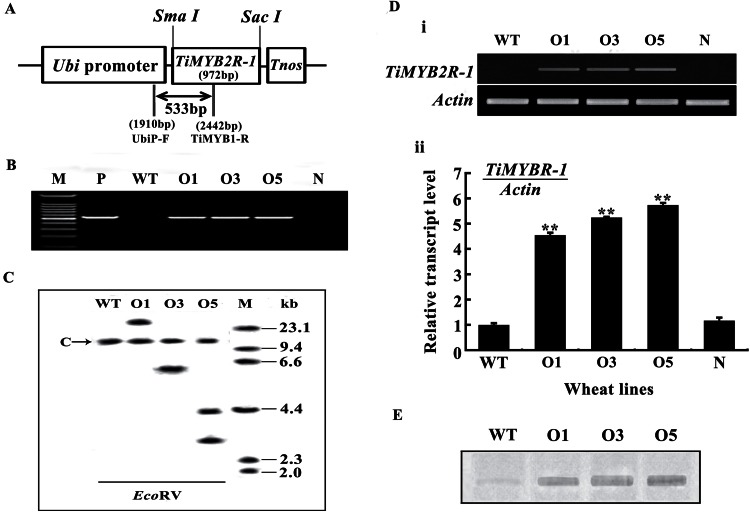
The presence of the introduced TiMYB2R-1 gene in transgenic wheat plants was monitored by PCR using Ubi::TiMYB2R-1-specific primers (Fig. 4A): UbiP-F, 5’-GCTCTGCCTTCATACGCTAT-3’ (located in the Ubi promoter of the transformation vector); and TiMYB1-R, 5’-TCCGCCAGTAGTTCTTGACC-3’ (located in the TiMYB2R-1 coding sequence). PCR was performed in a 25 µl volume containing 50ng of genomic DNA, 1× GC PCR buffer I (TaKaRa), 0.4 µM of each primer, 200 µM of each dNTP, and 1U of Taq polymerase (TaKaRa). The amplified product (533bp, Fig. 4A, B) specific to the Ubi::TiMYB2R-1 transgene was resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
Fig. 4.
TiMYB2R-1 transformation vector and molecular characterization of transgenic wheat. (A) In transformation vector pA25-TiMYB2R-1, the TiMYB2R-1 gene was driven by the maize ubiquitin (Ubi) promoter and terminated by Tnos. The arrow indicates the region amplified in the PCR assays using UbiP-F and TiMYB1-R primers and used for the Southern blot probe. (B) PCR pattern of TiMYB2R-1 transgenic and wild-type wheat plants using the transgene-specific primers UbiP-F and TiMYB1-R. M, 100bp DNA ladder; P, transformed vector plasmid pA25-TiMYB2R-1. (C) A Southern blot analysis of EcoRV-digested genomic DNAs from non-transformed and TiMYB2R-1 transgenic plants hybridized with an amplified fragment specific for the transgene. M, λDNA/HindIII markers. C band indicates the common bands in all wheat plants. (D) RT–PCR (i) and Q-RT–PCR (ii) analyses of TiMYB2R-1 transcript levels in the roots of T5 transgenics and control plants. In Q-RT–PCR analysis, three replicates for each sample were averaged, with the standard error of the mean (SE) indicated. Asterisks indicate statistically significant variation (**P < 0.01). (E) Western blot pattern of TiMYB2R-1 protein expression in roots of Ggt-resistant T5 transgenics and susceptible wild-type wheat lines. WT, non-transformed wheat Yangmai 12; O1, O3, and O5, three TiMYB2R-1 transgenic lines; N, segregant plants lacking TiMYB2R-1.
Southern blot analysis
Southern blotting was conducted following a modified protocol of Sharp et al. (1988). Genomic DNAs (~20 µg each) of the transgenic wheat plants and non-transformed Yangmai 12 were digested separately by the restriction enzyme EcoRV, and then blotted onto a Hybond N+ nylon membrane (GE Amersham). The amplified fragment (533bp, Fig. 4A) specific to the chimeric Ubi::TiMYB2R-1 was labelled by [α-32P]dCTP and then used as the probe. Hybridization was performed at 65 °C for 20h. The hybridized membrane was washed twice in 1× SSC, 0.1% SDS, twice in 0.5× SSC, 0.1% SDS, and once in 0.2× SSC, 0.1% SDS at 65 °C for 10min each time.
Transcription analyses of TiMYB2R-1 and wheat defence-related genes in transgenic wheat
In a previous microarray analysis, wheat TaPIMP1 overexpression activated transcription of defence-related genes (Zhang et al., 2012), including PR1a (TC384382), PR17c (TC398605), Chitinase 2 (Chit2; TC426538), class III chitinase (Chit3; TC386649), nsLTP1 (TC411506) encoding a non-specific lipid transfer protein1, and GST22 (TC372250). These TC numbers and sequences can be found at the website: http://compbio.dfci.harvard.edu/cgi-bin/tgi/tc_ann.pl?gudb=wheat.
Reverse transcription–PCR (RT–PCR) and real-time quantitative RT–PCR (Q-RT–PCR) were used to analyse transcription levels of TiMYB2R-1 and the above-mentioned wheat defence-related genes in TiMYB2R-1 transgenic and control wheat lines. The RT–PCRs for TiMYB2R-1 transcription were set up with 31 cycles (95 °C for 1min, 56 °C for 40 s, 72 °C for 35 s) to illustrate high expression of TiMYB2R-1 in the transgenics with primers (MRT-F and MRT-R) specific to the TiMYB2R-1 sequence (Table 1); the third from last nucleotide of the reverse primer MRT-R is different from that of TaPIMP1 (Supplementary Fig. S1 available at JXB online). Q-RT–PCR was performed using SYBR Green I Master Mix (TaKaRa, Japan) in a volume of 25 µl on an ABI 7300 RT–PCR system (Applied Biosystems). Reactions were set up with the following thermal profile: 95 °C for 5min, followed by 41 cycles of 95 °C for 15 s and 60°C for 35 s. The wheat actin gene was used to normalize amounts of cDNAs among the samples. The relative transcript level of a target gene was calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). The relative transcript levels of the tested genes in the transgenics were relative to those in the wild-type (WT) recipient since the genetic backgrounds of the transgenics and the WT recipient are the same except for the introduced TiMYB2R-1 and the reactions (including defence genes) caused by TiMYB2R-1 overexpression. In Q-RT–PCR analysis, transcription of TaPIMP1, a wheat endogenous homologue of TiMYB2R-1, may be detected in TiMYB2R-1 transgenics and WT recipient wheat. The relative transcript levels of TiMYB2R-1 were the transcript levels of TiMYB2R-1 and TaPIMP1 in transgenics minus those of TaPIMP1 in WT plants. Three replications for each treatment were performed.
Table 1.
Primers used in (Q-)RT–PCR.
| Gene name | Accession no. | MBS position and sequence | Sequence of gene-specific primer | Species |
|---|---|---|---|---|
| TiMYB2R-1 | JX683795 | F: 5’-ACGGACAACGAGGTCAAGAAC-3’ | Thinopyrum intermedium | |
| R: 5’-GAAATGGGCTCCGTACG-3’ | ||||
| PR1a | TC384382 | –43 to –39, GGATA | F: 5’-CGTGGGTGTCGGAGAAGC-3’ | Triticum aestivum |
| R: 5’-AAGTTGCCTGGCGGGTTG-3’ | ||||
| PR17c | TA65181 | –58 to –54 GGATA | F: 5’-ACGACATCACGGCGAGGT-3’ | Triticum aestivum |
| R: 5’-CACGGGGAAAGAGAGGATGA-3’ | ||||
| nsLTP1 | TC411506 | –86 to –81(–), CTGTTA | F: 5’-ATGCGGGTTGGCGTGAAG-3’ | Triticum aestivum |
| R: 5’-TGTTGCGGTGGTAGGTTGTTG-3’ | ||||
| GST22 | TC372250 | –211 to –206(–), CCGTTG | F: 5’-GGGATTGGGGCAGGAG-3’ | Triticum aestivum |
| R: 5’-TCGGGAGGGAGGAAGC-3’ | ||||
| Chit2 | TC426538 | No promoter sequence | F: 5’-TTCTGGATGACGGCACAAG-3’ | Triticum aestivum |
| R: 5’-CCTTAGTGTGACCAGTCGTTTT-3’ | ||||
| Chit3 | TC386649 | No promoter sequence | F: 5’-TAAGATGAGCCCCTACCTGTTC-3’ | Triticum aestivum |
| R: 5’-GTACACGGCATTTATTTAGTCCC-3’ | ||||
| Actin | BE425627 | F: 5’-CACTGGAATGGTCAAGGCTG-3’ | Triticum aestivum | |
| R: 5’-CTCCATGTCATCCCAGTTG-3’ | ||||
| 18SrRNA | AY049040 | F: 5’-GTGACGGGTGACGGAGAATT-3’ | Triticum aestivum | |
| R: 5’-GACACTAATGCGCCCGGTAT-3’ | ||||
| Ggt 18SrRNA | FJ771002 | F: 5’-CGAACTCGGTCGTTTAGAGG-3’ | Gaeumannomyces graminis var. tritici | |
| R: 5’-GGTATGTTCACAGGGGTTGG-3’ |
The primer sequences for TiMYB2R-1, and wheat defence-related and actin genes, and related information on (Q)-RT–PCR analyses are listed in Table 1.
Western blotting assay
The expression of the TiMYB2R-1-encoded protein in the transgenic wheat lines was tested by western blotting analysis. Total proteins were extracted from 0.3g of ground root powder. About 12 µg of total soluble proteins for each line were separated on 12% SDS–polyacrylamide gels and transferred to a polyvinyl difluoride membrane (Amersham). The western blots were incubated with the polyclonal GST–TiMYB2R-1 antibody (1:100 dilution), which was developed in mice from GST–TiMYB2R-1 protein. TiMYB2R-1 protein in these lines was visualized with the ECL Western Blot Detection and Analysis System (GE Healthcare).
Take-all assessments in transgenic wheat
Take-all responses of TiMYB2R-1 transgenics in the T3–T5 generations and untransformed wheat controls were evaluated following inoculation with Ggt. At 3 weeks post-inoculation, the disease severity in each plant was assessed as the percentage area of take-all lesions covering the root system (Daval et al., 2011). The infection types (ITs) were categorized from 0 to 4 according to Bithell et al. (2011) (i.e. IT 0, no take-all; IT 1, >0% and ≤10%; IT 2, >10% and ≤30%; IT 3, >30% and ≤60%; IT 4, >60%). The take-all index (TAI) was (10×N1+30×N2+60×N3+100×N4)/(N1+N2+N3+N4), where N was the number of plants with each infection type (Bithell et al., 2011). At least 70 plants per line were tested.
To investigate the take-all resistance of transgenic wheat lines further, Q-RT–PCR was used to assess the relative abundance of Ggt in transgenic wheat plants based on Ggt 18S rRNA (FJ771002; Daval et al., 2011) in reference to wheat 18S rRNA (AY049040). The primer sequences for Ggt 18S rRNA and wheat 18S rRNA in Q-RT–PCR analyses are listed in Table 1.
Results
TiMYB2R-1 is an R2R3-MYB transcription factor in T. intermedium
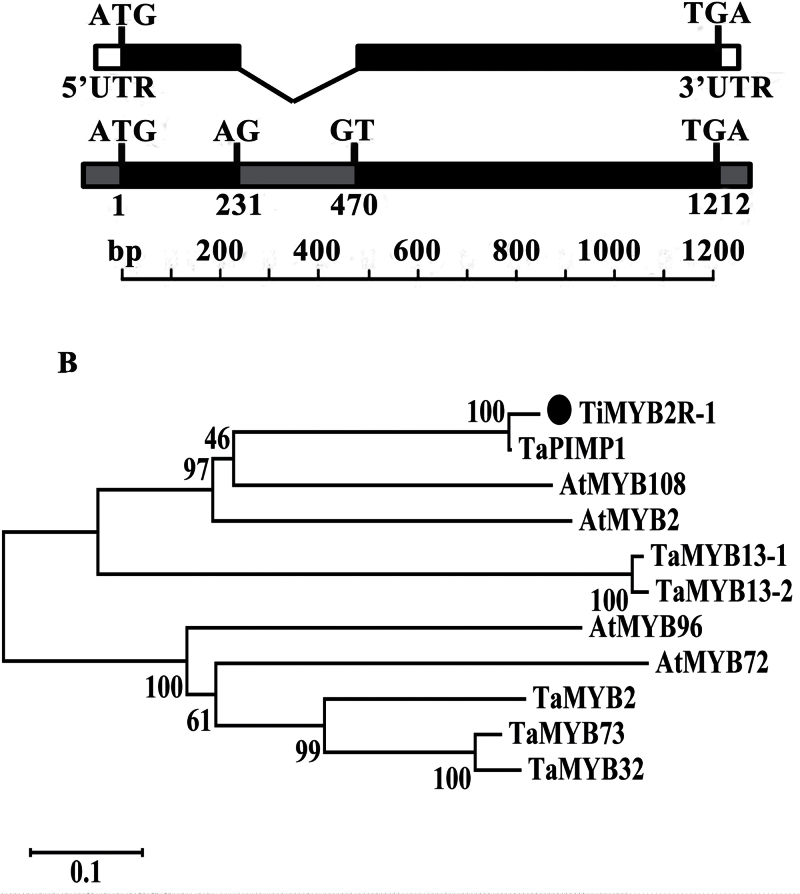
Both the full-length cDNA and genomic sequences of the TiMYB2R-1 gene were isolated from T. intermedium. The gene sequence includes two exons and an intron of 240bp (Fig. 1A). The ORF of TiMYB2R-1 encodes a deduced protein, TiMYB2R-1, with 323 amino acids and a mol. wt of 35.4kDa. The TiMYB2R-1 protein possesses two conserved SANT domains [one (R2) located at amino acids 46–94 and the other (R3) at amino acids 100–145], two acidic transcriptional activation domains (at amino acids 15–60 and 288–307), and two basic nuclear localization signal (NLS) regions (at amino acids 80–89 and 142–150) (Supplementary Fig. S2 at JXB online).
Fig. 1.
The gene structure and phylogenetic tree of TiMYB2R-1. (A) The TiMYB2R-1 predicted mRNA structure is shown above the genomic sequence with introns (grey), exons (black), and untranslated regions (white). ATG represents the start methionine codon and TGA represents the stop codon. (B) Phylogenetic tree constructed by Neighbor–Joining algorithms of MEGA 5.05 software after the multiple MYB protein sequence alignment using the CLUSTAL W program. Accession numbers for the other MYB proteins are: TaPIMP1 (ABU93236.1), TaMYB73 (AEW23186.1), TaMYB32 (AEV91155.1), TaMYB13-2 (AER38258.1), TaMYB13-1 (AER38255.1), TaMYB2 (AAT37168.1), AtMYB108 (AEE74402.1), AtMYB2 (BAA03534.1), AtMYB72 (AEE33352.1), and AtMYB96 (AED97611.1).
The overall sequence of the TiMYB2R-1 protein displayed various identities with known R2R3-MYB proteins from diverse species, namely wheat MYB proteins, including TaPIMP1 (ABU93236.1, 95.99%), TaMYB73 (AEW23186.1, 30.03%), TaMYB32 (AEV91155.1, 28.4%), TaMYB13-1 (AER38255.1, 27.24%), and TaMYB2 (AAT37168.1, 26.99%), and Arabidopsis MYB TFs, including AtMYB108 (AEE74402.1, 41.26%), AtMYB2 (BAA03534.1, 38.58%), AtMYB72 (AEE33352.1, 22.87%), and AtMYB96 (AED97611.1, 22.6%). Phylogenetic analysis indicated that TiMYB2R-1 clustered with R2R3-MYB proteins (Fig. 1B), among which AtMYB108, AtMYB96, and TaPIMP1 are implicated in responses to biotic and abiotic stresses in Arabidopsis and wheat (Mengiste et al., 2003; Seo and Park, 2010; Zhang et al., 2012). The above results suggest that TiMYB2R-1 is probably an R2R3-MYB protein.
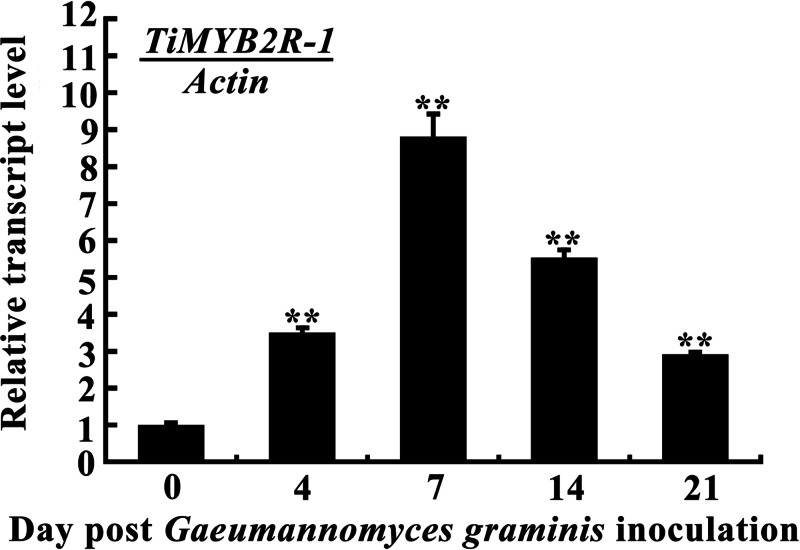
TiMYB2R-1 transcript level increases in T. intermedium after Ggt infection
Following Ggt inoculation, the transcription of TiMYB2R-1 in T. intermedium was significantly induced. The transcription level at 7 dpi reached a peak (8-fold above that of non-treated plants, 0h), declined afterwards, but remained higher than the non-treatment control (Fig. 2). The results suggested that TiMYB2R-1 is potentially involved in the host defence response to Ggt.
Fig. 2.
Transcription of TiMYB2R-1 in T. intermedium following Ggt challenge. The transcription level of TiMYB2R-1 in T. intermedium was measured by Q-RT–PCR relative to an untreated control (0h). Three biological replicates for each time point were averaged, with the standard error of the mean (SE) indicated. Asterisks indicate statistically significant variation calculated using the Student’s t-test (**P < 0.01).
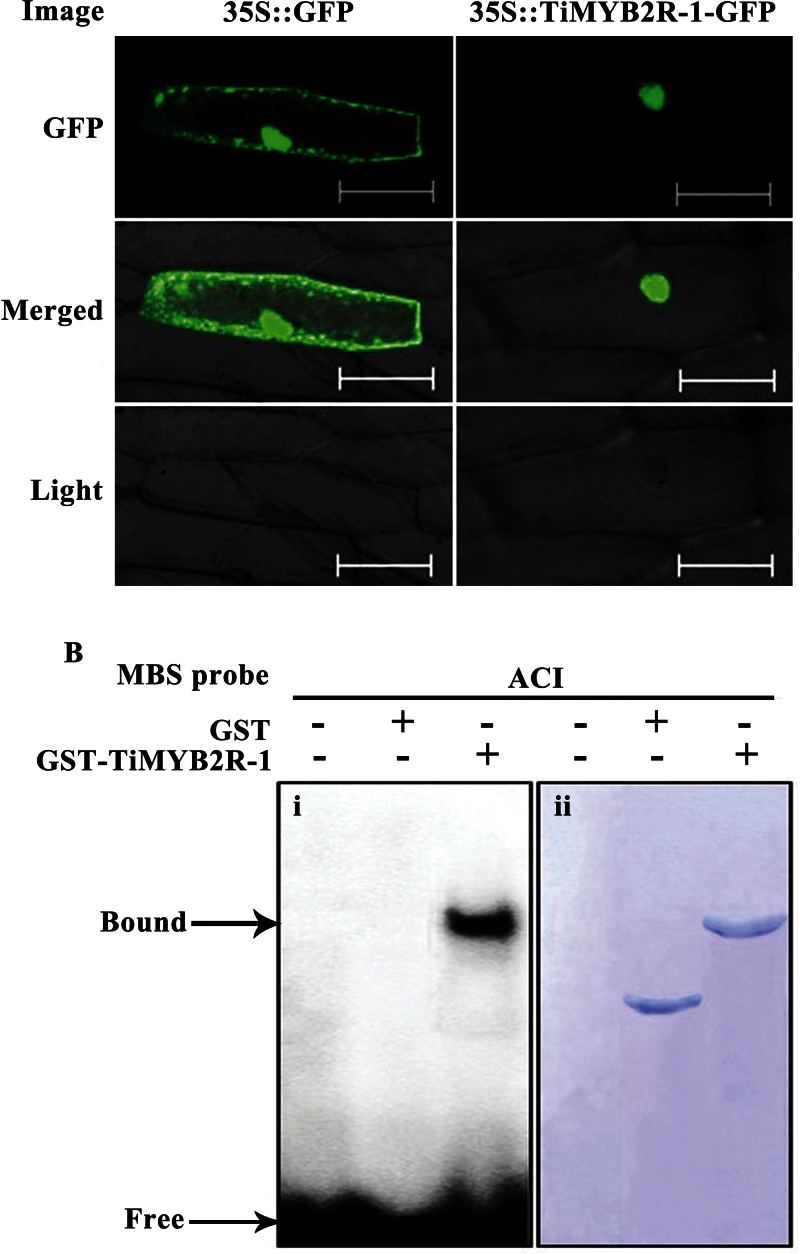
TiMYB2R-1 localizes to nuclei and binds to the MBS ACI cis-element
To study the subcellular localization of TiMYB2R-1, a chimeric p35S::TiMYB2R-1-GFP expression vector or control vector p35S::GFP was introduced separately into onion epidermal cells. Confocal imaging of transient expression in the epidermal cells showed that TiMYB2R-1–GFP accumulated only in the nucleus, whereas GFP alone was present throughout the whole cell, indicating that TiMYB2R-1 is a nuclear-localized protein (Fig. 3A). These results were consistent with those of TFs that typically function in the nuclei.
To test whether this TiMYB2R-1 protein bind to MBS ACI cis-elements, purified GST–TiMYB2R-1 recombinant protein was used in EMSAs. The results proved that the GST–TiMYB2R-1 could bind to the ACI cis-element, whereas GST protein alone did not (Fig. 3B). Thus TiMYB2R-1 is indeed an R2R3-MYB TF.
Fig. 3.
The subcellular localization and electrophoretic mobility shift assay (EMSA) of TiMYB2R-1 protein. (A) Subcellular localization of 35S::TiMYB2R-1–GFP fused protein in onion epidermal cells. p35S::TiMYB2R-1-GFP and p35S::GFP constructs were introduced separately into onion cells by bombardment, expressed, and observed under a confocal microscope. Bars=100 µM. (B) EMSA of TiMYB2R-1. Lane 1, only the ACI probe; lane 2, GST alone with the ACI probe; lane 3, GST–TiMYB2R-1 fusion protein with the ACI probe. (i) EMSA of DNAs in the gel stained by ethidium bromide. The free ACI probe and the retarded band of GST–TiMYB2R-1 fusion protein binding to the ACI probe are indicated. (ii) The proteins in the gel stained by Coomassie colloidal blue after EMSA. (This figure is available in colour at JXB online.)
Generation and molecular characterization of TiMYB2R-1 transgenic wheat
To evaluate the role of TiMYB2R-1 in wheat, transgenic wheat lines expressing TiMYB2R-1 were generated via bombarding the TiMYB2R-1 expression vector pA25-TiMYB2R-1 (Fig. 4A) into 1200 immature Yangmai 12 embryos. Four independent transgenic lines containing the Ubi::TiMYB2R-1 transgene were obtained with a transformation efficiency of 0.33%. Using the primers specific for the chimeric Ubi::TiMYB2R-1 (Fig.4A), genomic PCR assays of the T0–T5 generation plants showed that the specific band of the introduced TiMYB2R-1 was detected in progeny derived from three transgenic wheat lines O1, O3, and O5 with enhanced resistance to take-all, but not in non-transformed (WT) Yangmai 12 (recipient) and null-segregants lacking the transgene (Fig. 4B). Southern blot analysis of T4 transgenic wheat lines indicated that the three transgenic lines O1, O3, and O5 had one, one, and two copies, respectively, with different hybridization patterns (Fig. 4C), suggesting that the threee lines resulted from independent transformation events. Additionally, a common band was present in these transgenic lines and WT Yangmai 12, suggesting that the transgenic lines and Yangmai 12 contain a sequence (TaPIMP1) homologous to the probe derived from the Ubi::TiMYB2R-1 chimera (Fig.4A, C). Collectively these results proved that the transgene was integrated into the genomes of the three transgenic wheat lines and was stably transmitted to subsequent generations.
The expression of TiMYB2R-1 was examined in roots of three transgenic lines (O1, O3, and O5) with enhanced resistance at 21 d after Ggt inoculation. The RT–PCRs for TiMYB2R-1 transcription were set up with 31 amplified cycles to illustrate the higher expression of TiMYB2R-1 in the three transgenic lines, while no corresponding products were observed in WT Yangmai 12 and null-transgenic lines (Fig. 4Di). In Q-RT–PCR analysis, as TaPIMP1 transcription should be amplified at the same level in the transgenic and WT recipient plants, the relative transcript levels of TiMYB2R-1 were the transcript levels of TiMYB2R-1 and TaPIMP1 in transgenics relative to that of TaPIMP1 in the WT (Fig. 4Dii). As shown in Fig. 4Dii, TiMYB2R-1 in the transgenic lines was transcribed at a significantly higher level. Western blot results indicated that these lines (O1, O3, and O5) expressed high levels of TiMYB2R-1 protein, although the WT and transgenic lines contained a tiny amount of TaPIMP1 (Fig. 4E). Thus, the introduced TiMYB2R-1 gene can be highly expressed in the three transgenic lines with enhanced resistance.
Expression of TiMYB2R-1 improves resistance to take-all in transgenic wheat
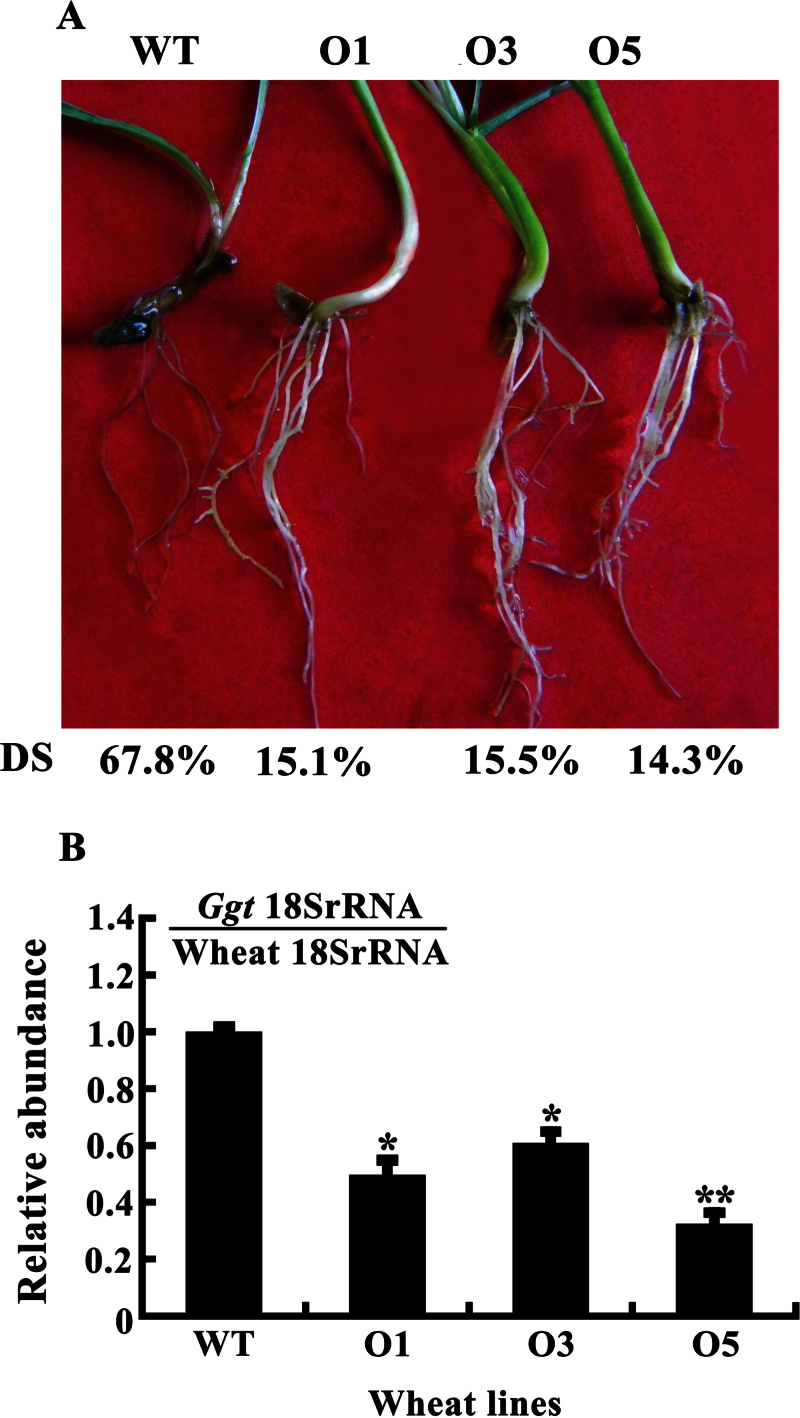
Take-all reactions of transgenic wheat lines overexpressing TiMYB2R-1 along with the null-segregants and WT Yangmai 12 were evaluated 21 d after Ggt inoculation. The results showed that most plants of TiMYB2R-1-overexpressing transgenic wheat lines O1, O3, and O5 displayed significantly increased resistance relative to the WT Yangmai 12 and null-segregant plants (Table 2, Fig. 5A). The average disease severities of the three transgenic wheat lines were 9.32–25.84%, whereas those of the null-segregants and WT Yangmai 12 were 43.13–49.25% and 44.74–50.14%, respectively. The average TAIs of these transgenic wheat lines were 18.75–35.47, whereas those of the null-segregant and WT Yangmai 12 were 63.33–66.75 and 61.67–68.89, respectively (Table 2). Moreover, using Ggt 18S rRNA levels as an indicator of the fungal biomass and infection, the relative Ggt abundance was significantly lower in the three TiMYB2R-1-overexpressing transgenic lines than in WT Yangmai 12 and null-segregants (Fig. 5B), further supporting that TiMYB2R-1 transgenic wheat plants were more resistant to Ggt infection. Thus, TiMYB2R-1-overexpressing transgenic wheat exhibited significantly enhanced resistance to take-all.
Table 2.
Take-all responses of TiMYB2R-1 transgenic and control wheat lines.a
| Lines | T4 generation | T5 generation | ||
|---|---|---|---|---|
| Disease severity (%) | Take-all index | Disease severity (%) | Take-all index | |
| O1 | 20.495** | 28.5** | 23.99** | 35.47** |
| O3 | 22.26** | 32.33** | 25.84** | 34.75** |
| O5 | 9.32** | 18.75** | 19.24** | 28.12** |
| Null | 43.13 | 63.33 | 49.25 | 66.75 |
| Yangmai12 (recipient) | 44.74 | 61.67 | 50.14 | 68.89 |
Significant difference between TiMYB2R-1 transgenic lines and untransformated Yangmai 12 (recipient) at **P <0.01.
Null indicates the segregants lacking TiMYB2R-1.
a The values derived from the average of 70 plants of each line tested in T4 and T5 transgenics and control wheat lines.
Fig. 5.
Typical phenotypes of take-all and relative abundance of Ggt in TiMYB2R-1 transgenics and non-transformed wheat Yangmai 12 plants. (A) Responses of resistant TiMYB2R-1 transgenic wheat lines, and a susceptible plant of non-transformed Yangmai 12 to take-all. DS indicates the disease severity of take-all in wheat roots. (B) The relative abundance of Ggt in resistant transgenic wheat lines (O1, O3, and O5) relative to that in non-transformed Yangmai 12 (WT) based on Ggt 18S rRNA in reference to wheat 18S rRNA. Three replicates of each line were averaged, with the standard error of the mean (SE) indicated. Asterisks indicate statistically significant variation (**P < 0.01, *P < 0.05). (This figure is available in colour at JXB online.)
Expression of TiMYB2R-1 activates defence-related genes in transgenic wheat
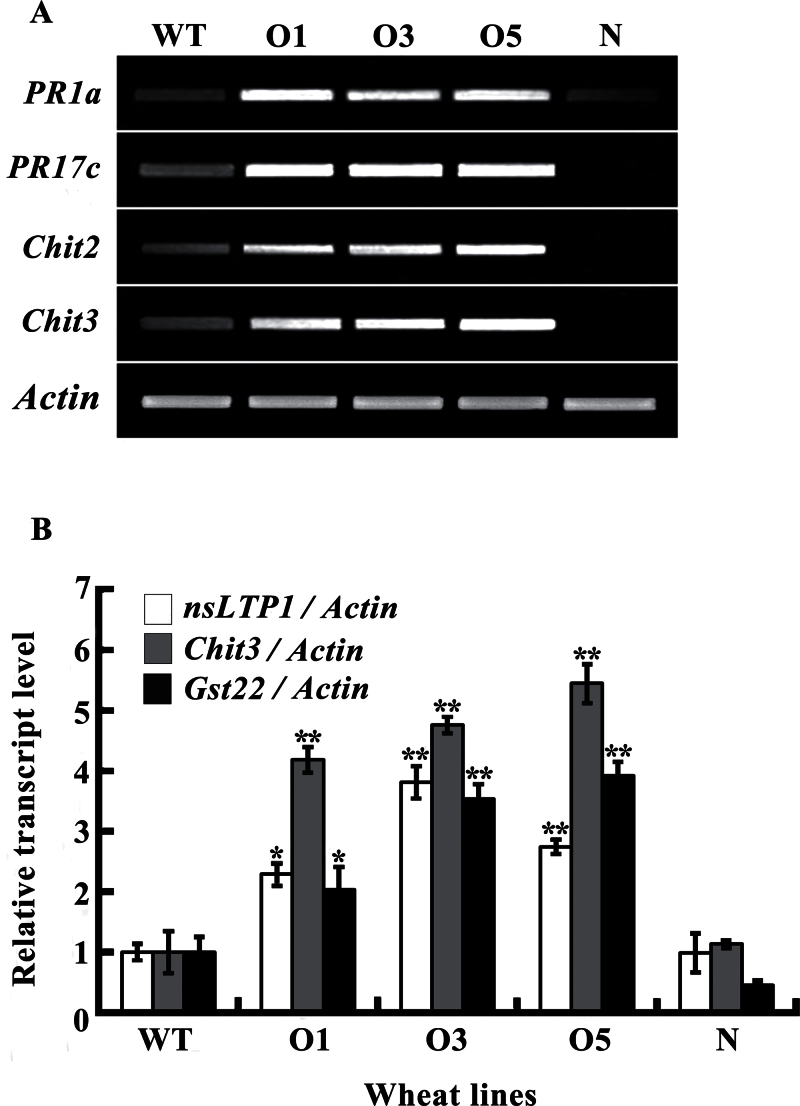
To investigate putative molecular mechanisms of TiMYB2R-1 overexpression in enhanced disease resistance, RT–PCR and Q-RT–PCR were used to analyse the transcription levels of six defence-related genes, namely PR1a, PR17c, Chit2, Chit3, nsLTP1, and GST22, in roots of TiMYB2R-1-overexpressing transgenics and of WT Yangmai 12 and null-segregant plants at 21 dpi with Ggt. As shown in Fig. 6, the transcription levels of all six genes were markedly elevated in TiMYB2R-1-overexpressing lines relative to those in the null-segregant and WT plants. The results suggested that overexpression of TiMYB2R-1 activates transcription of these defence-related genes in the transgenic wheat lines.
Fig. 6.
Expression patterns of the defence-related genes PR1a, PR17c, Chit2, Chit3, nsLTP1, and GST22 in roots of Ggt-resistant T5 transgenics and WT and null-segregant plants. (A) RT–PCR analyses. (B) Q-RT–PCR analyses. O1, O3, and O5, three TiMYB2R-1 transgenic lines; N, null-segregants lacking TiMYB2R-1; WT, non-transformed Yangmai 12. The transcript level of a defence-related gene in resistant transgenics (O1, O3, and O5) was relative to that in WT Yangmai 12. Three replicates for each sample were averaged, with the standard error of the mean (SE) indicated. Asterisks indicate statistically significant variation (**P < 0.01, *P < 0.05).
Discussion
In plant species, some MYB proteins have been implicated in defence responses (Mengiste et al., 2003; Seo and Park, 2010; Zhang et al., 2012). Although T. intermedium possesses resistance to diverse pathogens, it is not known if and how MYB TFs in T. intermedium regulate defence responses to the pathogens. In this study, the first MYB gene of T. intermedium, namely TiMYB2R-1, was successfully isolated. Sequence analysis showed that the gene sequence of TiMYB2R-1 includes two exons and an intron, and the deduced TiMYB2R-1 protein possesses the structural characteristics of R2R3-MYB TFs. Sequence alignment and phylogenetic analysis indicated that TiMYB2R-1 protein was most closely related to a wheat R2R3 MYB TaPIMP1, followed by Arabidopsis AtMYB108. The results of subcellular localization and MBS cis-element binding analyses proved that TiMYB2R-1 indeed is an R2R3-MYB TF in T. intermedium, consistent with its sequence traits.
Many TFs are induced under environmental stress conditions. For example, transcriptional levels of wheat TaMYB73 and Arabidopsis AtMYB44 were up-regulated under salinity stress conditions (He et al., 2011). The expression of the wheat MYB gene TaPIMP1 was increased after Bipolaris sorokiniana infection and dehydration treatment (Zhang et al., 2012). They can offer potential genes for improving biotic and abiotic stress in planta. The transcription of TiMYB2R-1 in T. intermedium was significantly induced after inoculation with Ggt or B. sorokiniana (Supplementary Fig. S3 at JXB online), suggesting that TiMYB2R-1 is probably involved in host response to infection by Ggt or B. sorokiniana.
To investigate defence roles of TiMYB2R-1 in planta, here TiMYB2R-1-overexpressing transgenic wheat lines were generated through transformation and characterized in detail. Based on PCR detection for T0–T5 transgenic wheat lines and Southern blot analyses using a transgene-specific amplicon as a probe, the results showed that the TiMYB2R-1 transgene was integrated into the genomes of three transgenic wheat lines and could be transmitted to subsequent generations. RT–PCR and western blot assays indicated that TiMYB2R-1 was highly expressed in the three transgenics. Following inoculation with the take-all pathogen Ggt, the disease severity, TAI, and Ggt relative biomass assays showed that the transgenic wheat lines overexpressing TiMYB2R-1 had more significantly enhanced resistance to take-all than WT Yangmai 12 and null-segregants, although TaPIMP1 exists in the WT and TiMYB2R-1 transgenic wheat lines and is induced following Ggt infection. The enhanced degrees of resistance in the transgenic lines were correlated with TiMYB2R-1 expression levels. These suggested that overexpression of TiMYB2R-1 conferred increased resistance in transgenic wheat, whereas the expression of TaPIMP1 in WT Yangmai 12 is not enough to confer resistance after Ggt challenge. Take-all responses of TaPIMP1-overexpressing wheat will be tested in the future. Furthermore, overexpression of TiMYB2R-1 does not affect the development and growth of the transgenic wheat under normal growth conditions. Thus, TiMYB2R-1 can be used as an important engineering gene for improving take-all resistance of wheat. Additionally, the transgenic wheat lines expressing TiMYB2R-1 showed enhanced resistance to common root rot caused by B. sorokiniana (Supplementary Table S1 at JXB online), similar to TaPIMP1-overexpressing wheat (Zhang et al., 2012). Thus, the transgenic wheat produced in this study will provide potential wheat germplasm for enhancing resistance to take-all and common root rot. This is believed to be the first report on development of take-all-resistant wheat lines.
In plants, certain activator-type TFs have been implicated in triggering disease resistance and abiotic stress tolerance through activation of defence- and stress-related genes (Ma et al., 2009; Zhang et al., 2012). Defence-related genes play vital roles in defence against pathogens in plants (Guilleroux and Osbourn, 2004; Seo and Park, 2010; Zhang et al., 2012). For example, transgenic wheat plants overexpressing a wheat LTP gene or a barley chitinase gene displayed increased resistance to fungal pathogens (Shin et al., 2008; Zhu et al., 2012). Although no natural highly resistant wheat cultivar has been identified, the transcription levels of some wheat defence-responsive genes were up-regulated during compatible interactions between wheat roots and Ggt using suppression subtractive hybridization and expressed sequence tag (EST) analysis (Guilleroux and Osbourn, 2004). Among them, two ESTs corresponded to Chit3 and GST2 genes (Guilleroux and Osbourn, 2004). TFs interact with specific DNA sequences (cis-acting elements) in target genes to modulate the transcription process. In a previous study, the microarray data showed that wheat TaPIMP1 overexpression activates the transcription of some defence-related genes in wheat, namely PR1a, PR17c, Chit2, Chit3, nsLTP1, and GST22 (Zhang et al., 2012).
In this study, RT–PCR and Q-RT–PCR were used to investigate if TiMYB2R-1 overexpression activates the same defence-related genes in the transgenic wheat, using the null-segregant and WT wheat lines as controls. The results indicated that overexpression of TiMYB2R-1 indeed elevated the expression of such defence-related genes in the transgenic wheat. The promoter sequences of these defence-related genes obtained from the Wheat Draft Genome Assembly database (http://www.cerealsdb.uk.net/CerealsDB/), and prediction of MBS cis-elements in the promoters (www.dna.affrc.go.jp/PLACE/) revealed that MBS cis-acting elements exist in the promoters of wheat defence genes PR1a, PR17c, nsLTP1, and GST22 (Table 1). TiMYB2R-1 possibly interacts with these promoters and thereby contributes to the increased expression of these genes. The promoters of wheat Chit2 and Chit3 genes could not be found due to the incomplete coverage of wheat genome sequences. TiMYB2R-1 may elevate the expression of Chit2 and Chit3 genes through indirect means, which has yet to be verified experimentally. The results suggested that changes in expression of a subset of wheat defence-related genes regulated by TiMYB2R-1 expression probably result in enhanced resistance to take-all in TiMYB2R-1 transgenic wheat. The results may provide a new insight into the interaction between wheat and Ggt.
In summary, TiMYB2R-1, the first MYB gene isolated from T. intermedium, was characterized. It encodes an R2R3-MYB TF and has significantly higher expression levels in host plants following Ggt infection. TiMYB2R-1 overexpression in transgenic wheat lines showed significantly enhanced resistance to take-all and common root rot possibly through activation of some defence-related genes. The results contribute to further understanding of the characteristics and functions of the MYB TF family in additional plant species.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Alignment of TiMYB2R-1 and TaPIMP1 sequences amplified in (Q-)RT–PCR.
Figure S2. The cDNA sequence and deduced amino acid sequence of TiMYB2R-1.
Figure S3. Transcription analysis of TiMYB2R-1 in T. intermedium following B. sorokiniana inoculation.
Table S1. Bipolaris sorokiniana responses of TiMYB2R-1 transgenic and control wheat.
Acknowledgements
The authors are grateful to Dr Cunjin Zhang (Durham University, UK) for revision, and Professor Daowen Wang (Chinese Academy of Sciences) for providing the ph16318 vector. This study was supported by the National ‘863’ project (grant no. 2012AA10A309), a National ‘Key Sci-Tech’ projects (2011ZX08002-001 and 2013ZX08002-001), and the NSFC programme (grant no. 30871523).
References
- Banks PM, Larkin PJ, Bariana HS, et al. 1995. The use of cell culture for subchromosomal introgressions of barley yellow dwarf virus resistance from Thinopyrum intermedium to wheat. Genome 38, 395–405. [DOI] [PubMed] [Google Scholar]
- Bithell SL, Butler RC, Harrow S, Mckay A, Cromey MG. 2011. Susceptibility to take-all of cereal and grass species, and their effects on pathogen inoculum. Annals of Applied Biology 159, 252–266. [Google Scholar]
- Cauderon Y, Saigne B, Dauge M. 1973. The resistance to wheat rusts of Agropyron intermedium and its use in wheat improvement. In: Sears ER, Sears LMS, eds. Proceedings of the 4th International Wheat Genet Symposium. University of Missouri, Columbia, MO, 401–407. [Google Scholar]
- Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wendel JF. 2003. Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Molecular Biology 51, 313–325. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang Z, Liang H, Liu H, Du L, Xu H, Xin Z. 2008. Overexpression of TiERF 1 enhances resistance to sharp eyespot in transgenic wheat. Journal of Experimental Botany 59, 4195–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang X, He K, et al. 2006. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology 60, 107–124. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. 1996. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Cook RJ. 2003. Take-all of wheat. Physiological and Molecular Plant Pathology 62, 73–86. [Google Scholar]
- Daval S, Lebreton L, Gazengel K, Boutin M, Guillerm-Erckelboudt A, Sarniguet A. 2011. The biocontrol bacterium Pseudomonas fluorescens Pf29Arp strain affects the pathogenesis-related gene expression of the take-all fungus Gaeumannomyces graminis var. tritici on wheat roots. Molecular Plant Pathology 12, 839–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E. 2003. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiology 131, 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis . Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Fedak G, Han F. 2005. Characterization of derivatives from wheat–Thinopyrum wide crosses. Cytogenetic and Genome Research 109, 360–367. [DOI] [PubMed] [Google Scholar]
- Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ashwin G, Parida A. 2012. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. Journal of Experimental Botany 63, 4549–4561. [DOI] [PubMed] [Google Scholar]
- Guilleroux M, Osbourn A. 2004. Gene expression during infection of wheat roots by the ‘take-all’ fungus Gaeumannomyces graminis . Molecular Plant Pathology 5, 203–216. [DOI] [PubMed] [Google Scholar]
- Gutteridge RJ, Bateman GL, Todd AD. 2003. Variation in the effects of take-all disease on grain yield and quality of winter cereals in field experiments. Pest Management Science 59, 215–224. [DOI] [PubMed] [Google Scholar]
- He Y, Li W, Lv J, Jia Y, Wang M, Xia G. 2011. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana . Journal of Experimental Botany 63, 1511–1522. [DOI] [PubMed] [Google Scholar]
- Li H, Arterburn M, Jones S, Murray T. 2005. Resistance to eyespot of wheat, caused by Tapesia yallundae, derived from Thinopyrum intermedium homoeologous group 4 chromosome. Theoretical and Applied Genetics 111, 932–940. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhou X, Dong N, Liu X, Zhang H, Zhang Z. 2011. Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Functional and Integrative Genomics 11, 431–443. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, et al. 2009. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology 150, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. 2003. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis . The Plant Cell 15, 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, Mclnnis S, Courtenay A, et al. 2003. Characterisation of a pine MYB that regulates lignification. The Plant Journal 36, 743–754. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO Journal 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Wang M, Tian Y, He W, Han L, Xia G. 2012. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis . Molecular Biology Reports 39, 7183–7192. [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E. 1999. Maize R2R3 Myb genes: sequence analysis reveals amplification in the higher plants. Genetics 153, 427–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JM, Dubos C, Prouse MB, et al. 2012. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytologist 195, 774–786. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM. 2010. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis . New Phytologist 186, 471–483. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. 2009. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiology 151, 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma HC, Gill BS, Uyemoto JK. 1984. High levels of resistance in Agropyron species to barley yellow dwarf virus and wheat streak mosaic viruses. Journal of Phytopathology 110, 143–147. [Google Scholar]
- Sharma H, Ohm H, Goulart L, Lister R, Appels R, Benlhabib O. 1995. Introgression and characterization of barley yellow dwarf virus resistance from Thinopyrum intermedium into wheat. Genome 38, 406–413. [DOI] [PubMed] [Google Scholar]
- Sharp PJ, Kreis M, Shewry PR, Gale MD. 1988. Location of -amylase sequences in wheat and its relatives. Theoretical and Applied Genetics 75, 286–290. [Google Scholar]
- Shen H, He X, Poovaiah CR, et al. 2012. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytologist 193, 121–136. [DOI] [PubMed] [Google Scholar]
- Shin S, Mackintosh CA, Lewis J, Heinen SJ, Radmer L, Dill-Macky R, Baldridge GD, Zeyen RJ, Muehlbauer GJ. 2008. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. Journal of Experimental Botany 59, 2371–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Xu H, Chen X, et al. 1991. Development of common wheat germplasm resistant to barley yellow dwarf virus by biotechnology. Science in China (Series B) 34, 1055–1062. [Google Scholar]
- Xue G, Kooiker M, Drenth J, Mclntyre CL. 2011. TaMYB13 is a transcriptional activator of fructosyltransferase genes involved in -2,6-linked fructan synthesis in wheat. The Plant Journal 68, 857–870. [DOI] [PubMed] [Google Scholar]
- Yang M, Mavrodi DV, Mavrodi OV, Bonsall RF, Parejko JA, Paulitz TC, Thomashow LS, Yang HT, Weller DM, Guo JH. 2011. Biological control of take-all by fluorescent Pseudomonas spp. from Chinese wheat fields. Phytopathology 101, 1481–1491. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, Liu X, Kong X. 2011. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. Journal of Experimental Botany 63, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Wang X, Zhou M, Zhou X, Ye X, Wei X. 2012. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host-resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytologist 196, 1155–1170. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li Z, Xu H, Zhou M, Du L, Zhang Z. 2012. Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to both Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Functional and Integrative Genomics 12, 481–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.