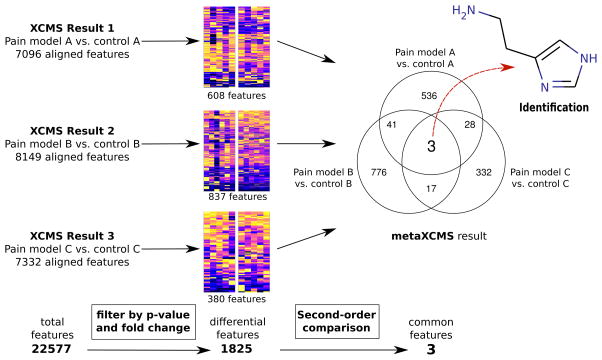
Figure 1.
Data reduction with metaXCMS. The workflow is demonstrated using the pain dataset, where pain model A is CFA-treated animals, pain model B is heat-treated animals, and pain model C is KRN-treated animals. The applied fold changes and p-values for the first filtering step were ≥ 1.5 and ≤ 0.05, respectively.