Fig. 3.
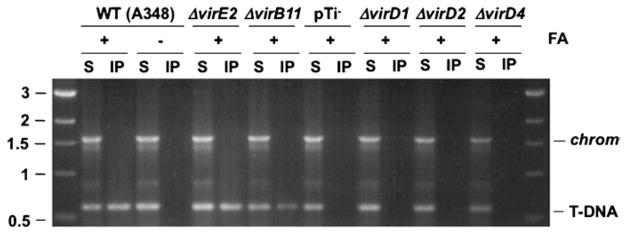
Isolation of substrate DNA–channel subunit complexes with the TrIP assay. The translocating T-DNA substrate is formaldehyde cross-linked to channel subunits as described in Subheading 3.6.1. The resulting complexes are recovered by immunoprecipitation with antisera against the channel subunits as described in Subheading 3.6.2. The precipitated DNA substrate is detected by PCR amplification as described in Subheading 3.6.3. Here, anti-VirD4 antisera was used to precipitate the VirD4 substrate receptor following FA-cross-linking and disruption of the strains shown. Total soluble (S) and immunoprecipitated (IP) material were assayed for the presence of substrate T-DNA (T-DNA) or the chvE gene (chrom) carried on the chromosome. Strains: WT strain A348; pTi-, A348 lacking the pTi plasmid; nonpolar deletion mutants - ΔvirE2 (virE2 codes for the exported effector VirE2; ΔvirD1 and ΔvirD2 (virD1 and virD2 code for relaxosomal subunits which are required for processing of the T-DNA substrate); ΔvirD4 (virD4 codes for the substrate receptor). A precipitable VirD4-T-DNA complex is recovered from FA-cross-linked WT cells and mutants lacking the VirE2 effector and VirB11 ATPase, but not from WT cells without prior FA cross-linking or mutants lacking the T-DNA processing factors VirD1 and VirD2 or the substrate receptor VirD4 (23).
