Abstract
In culture, exposure to penicillin and other stressors induce chlamydiae to enter a non-infectious but viable state termed persistence. Chlamydiae may reenter their normal developmental cycle after stressor removal. Though aberrant RB similar to those present in culture models of persistence have been observed within infected tissues, the existence of persistent chlamydiae has not been definitively demonstrated in vivo. As a result, the role of persistent organisms in pathogenesis is undefined. In order to establish an experimentally tractable model of in vivo persistence, C. muridarum vaginally-infected mice were gavaged with either water or amoxicillin (amox). Vaginal swabs were collected for chlamydial titration and RNA isolated for quantification of pre-16s rRNA. Uterine tissue was analyzed by transmission electron microscopy (TEM). Although amox-treatment reduced vaginal shedding by >99%, C. muridarum pre-16s rRNA accumulation was unchanged by treatment. These data indicate that the amox-exposed organisms were viable but not infectious. Furthermore, TEM analyses demonstrated that inclusions in amox-treated animals contained primarily large, aberrant RB, but those observed in untreated control animals were normal. Collectively, these data suggest that amoxicillin treatment induces C. muridarum to enter the persistent state in vivo. This model also represents the first experimentally tractable animal model of chlamydial persistence.
Keywords: C. trachomatis, C. muridarum, chlamydial persistence, aberrant reticulate body, aberrant body, amoxicillin
1. Introduction
Chlamydia trachomatis is the most common sexually transmitted bacterial disease agent worldwide. With more than 4 million new cases per year in the US, it is known as the “silent epidemic” because many patients are asymptomatic. Disease, however, can range from mild, asymptomatic infection to severe inflammation resulting in epididymitis, proctitis, pelvic inflammatory disease, infertility, and ectopic pregnancy.
C. trachomatis is a Gram-negative, obligate intracellular bacterium that exists in at least two morphologically distinct forms. The elementary body (EB, 0.2 um diameter) has a condensed genome and is able to infect a cell but is non-replicative. The reticulate body (RB, 0.8 um diameter) is able to replicate but is not infectious. After attachment to an epithelial cell surface, EBs are internalized by receptor-mediated endocytosis. Vacuoles containing EBs fuse to form an inclusion, an enlarged membrane-bound endosomal sac in which EBs transform to RBs. RBs acquire host cell metabolites for growth and divide by binary fission. After multiple rounds of division, RBs condense to form intermediate bodies (IB) and then infectious EBs, which are released and can infect new host cells [1].
In culture, environmental stressors such as cytokine (IFN-gamma) exposure, nutrient deprivation, and penicillin exposure can disrupt the normal developmental cycle by inducing the bacteria to enter a state variously termed persistence [1,2] or the chlamydial stress response [3]. Chlamydial persistence is defined as a developmental state in which the bacteria are viable but not infectious. Notably, persistent organisms can remain viable in culture for extended periods [4] and can reenter the normal developmental cycle after the persistence inducer is removed. Incredibly, Galasso and Manire demonstrated that penicillin-exposed C. psittaci could be maintained in culture for up to 9 months in this non-infectious but viable state, and that EB production ensued when the stressor was removed [5]. Subsequent transmission electron microscopy (TEM) analyses of penicillin G-exposed C. psittaci revealed that RB to EB conversion was inhibited, allowing the formation of enlarged, irregular, less electron dense developmental forms [6]. These enlarged aberrant bodies (AB), as well as reduced infectious EB production, are characteristic of the chlamydial persistent state [1,2].
Continued [7,8] and recurrent infections with the same serovar [9,10] occur in patients following the recommended treatment regimens. Chlamydial DNA, RNA, and antigens are also present in mice and non-human primates subsequent to culture-apparent resolution [11–13]. These observations have led some investigators to hypothesize that the persistent state may occur in vivo as well. Viable chlamydiae have been identified in patients with C. trachomatis-induced reactive arthritis [14] and chlamydiae showing morphologic characteristics of persistent organisms have been observed in patient samples [15,16]. More recently, Pospischil et al. convincingly demonstrated ABs in intestinal tissues of C. suis- infected swine [17] and Rank et al. found ABs in uterine samples from C. muridarum-infected mice [18]. Clinical signs of infection also recur in infected cats and mice treated with clavulanate-potentiated amoxicillin after drug therapy is terminated [19,20]. Though all of these data are consistent with persistence induction in vivo, they are limited by the fact that the investigators used either TEM or pre-16S rRNA analyses, but not both, in their studies. Thus, though multiple lines of evidence suggests that the “viable but non-infectious” state occurs in vivo, complete characterization of persistent organisms and the role they may play in chlamydial pathogenesis in vivo is yet to be determined.
Several investigators have hypothesized that persistent chlamydiae play a role in aiding chronic chlamydial infections and the resulting pathologies [21,22]. Unfortunately, lack of a tractable animal model of chlamydial persistence has hampered study of the effects of persistence in vivo. Most importantly, an in vivo model should allow the investigator to turn persistence on and off in a controlled manner. It is also important that the persistence inducer be non-toxic to the animal host. IFN-gamma is the most well characterized persistence inducer in culture [2] and its production can be knocked out in vivo. Unfortunately, there is no practical way to add IFN-gamma back into an animal without causing uncontrollable immunomodulatory effects. In vivo chlamydial persistence models based on nutrient starvation are equally unfeasible. Our recent studies indicate that amoxicillin (amox), the most commonly prescribed beta-lactam in the U.S., induces C. trachomatis persistence in culture. Here, we examine the suitability of amox for persistence induction in C. muridarum-infected mouse cells, and, for the first time, fully characterize chlamydial persistence and recovery in an inducible murine model.
2. Methods
2.1. Chlamydia, cells, and animals
C. muridarum Weiss strain was obtained from Dr. Kyle Ramsey and propagated in the Hec1B human endometrial cell line (ATCC# HTB-113) using bead culture [23]. Hec1B cells were cultured in Modified Essential Medium (MEM) with Hank's salts + 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Chlamydial stocks were titered in BM1.11 murine oviduct epithelial cells, generously provided by Dr. Ray Johnson [24], to determine total inclusion forming units (IFU)/mL. Vaginal swabs were titered on Hela 229 monolayers (ATCC# CCL2.1), which were grown in MEM with Earl's salts + 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Four to six week old female BALB/c mice (Harlan, USA) were allowed to acclimate to our animal facilities for one week, after which they were treated with 2.5 mg Depo-provera (Depo, Greenstone LLC, Peapack, NJ) by subcutaneous injection to synchronize their menstrual cycles and increase infection efficiency. One week after Depo treatment, mice were vaginally infected with 104-106 IFU of C. muridarum or an equal volume of 2SPG (0.2 M sucrose, 20 mM phosphate buffer, and 5 mM L-glutamine). Mice were sacrificed 21 days post infection (dpi).
2.2. BM1.11 cell infection, amox-exposure in vitro, and chlamydial titration by subpassage
BM1.11 cells were plated in 24 well plates at a density of 105 cells per well and either mock infected with 2SPG or experimentally infected with a dilution of C. muridarum inoculum sufficient to achieve ∼90% infectivity (∼1 MOI). After 1 h of adsorption, host cells were refed with 1:1 Dulbecco's modified Eagle medium:F12K (Sigma Chemical Co., St. Louis, Mo.) supplemented with 10% characterized FBS (HyClone, Logan, Utah), 2 mM L-alanyl-L-glutamine (Glutamax I; Gibco/Invitrogen, Carlsbad, Calif.), 5 ug of bovine insulin/ml, and 12.5 ng of recombinant human keratinocyte growth factor (KGF; Sigma Chemical Co.)/ml. Replicate cultures were either exposed to medium + diluent (ddH2O) or medium + amox at the time intervals described in section 2.3 below. At the indicated times, monolayers were scraped into 1 mL of culture medium and lysed by freeze/thaw and brief sonication. Cell lysates were centrifuged at 1000 × g for 5 min to pellet cell debris; the supernatants were then centrifuged at 8000 × g for 30 min to pellet chlamydial EBs. Chlamydial pellets were resuspended in 200 uL of 2SPG, diluted, and used to infect Hela cells plated at 1.5×105 cells per well on glass coverslips in triplicate. Infected monolayers were incubated for 24 h before being fixed and stained. Total inclusions per coverslip were counted and triplicate counts averaged. IFU/mL were then calculated and reported as IFU/mL +/- SEM. Significance was determined by Student's t-test with p<0.05 considered significant (*).
2.3. Amox exposure in vitro
BM1.11 cells were plated as described above and infected with C. muridarum Weiss. At 8 hours post infection (hpi), culture medium was replaced with amox-supplemented medium (0.605 mg/mL) or medium + diluent. Thirty hours after supplementation, samples were collected for titer and TEM, and the remaining replicate cultures were either refed again with amox-supplemented medium or culture medium + diluent. Cells were incubated for an additional 32 h to allow bacterial recovery from persistence before samples were again collected for titer and TEM.
2.4. Amox treatment in vivo
Mice were treated with 2 mg/kg or 40 mg/kg amox (Biomax, VIRBAC AH, Inc, Fort Worth, TX) in sterile water twice daily by gavage beginning at either 5 dpi or 3 dpi and continuing for 1 week. Control mice were gavaged with sterile water.
2.5. Quantification of chlamydial shedding
Mice were vaginally swabbed every third day with a calcium alginate swab (Fisher Scientific, Pittsburg, PA) to collect any infectious chlamydiae shed. Swabs were snap frozen in 2 mL tubes containing 500 uL of 2SPG and three 3 mm glass beads. Samples were later thawed, vortexed, and briefly sonicated to release EB from infected cells. Dilutions of the sample were used to infect Hela 229 cells plated at 105 cells per well on glass coverslips in 24 well plates. Cultures were centrifuged at 1100 × g for 1 h and refed with antibiotic/antifungal medium (MEM with 10%FBS, 1.77 uM cyclohexamide, 20 ng/mL gentamycin, 67.3 uM vancomycin, 1.4 uM fungizone, and 8.3 mM glucose). Chlamydial inclusions were allowed to develop for 24 h before being formaldehyde fixed and T×100 permeablized [25]. Inclusions were enumerated using Pathfinder anti-chlamydial LPS fluorescent stain (Bio-rad Laboratories, Hercules, CA) for visualization. Infectious shedding was reported as average IFU/mouse +/- SEM. Significance was determined by Student's paired t-test with p<0.05 considered significant (*).
2.6. Transmission electron microscopy (TEM)
Cultured cells were scraped into gluteraldehyde/paraformaldehyde TEM fixative and incubated at 4°C for 1 h before further processing. Mice were sacrificed at 6 or 7 dpi and the uterus carefully dissected from each animal to exclude any fatty tissue and to include as much of the cervix as possible. Collected tissues were refrigerated overnight in TEM fixative. All samples were washed with cacodylate buffer and then incubated with 1% osmium tetroxide in cacodylate buffer for 1 h (cell pellets) or 4 h (tissues) at room temperature. Samples were again washed with cacodylate buffer and dehydrated through a concentrated ethanol series before infiltration with Eponate 812 (Polysciences, Inc., Warrington, PA). Finally, samples were embedded in fresh Epon, thin sectioned, and visualized using a Tecnai Philips Transmission Electron Microscope at 80kV[18]. All electron micrographs were taken at 7000× magnification.
2.7. Polymerase chain reaction (PCR) and reverse-transcription (RT) PCR
Total RNA and DNA were isolated from day 3 and day 6 pi swab samples using the Quigen RNeasy mini column isolation kits (Qiagen, Inc., Maryland, USA). RNA integrity was determined by Agilent bioanalysis and equal volumes of samples with a RIN of 9 or higher were used to make cDNAs for quantification of chlamydial pre-16s ribosomal RNA following the protocol outlined by Deka et al [25]. These were normalized to chlamydial genome (16s rDNA) and host genome quantity (b-actin) [25]. Published primer sets include chlamydial 16s rRNA primary transcript [26], chlamydial 16s rRNA/gDNA [25], and human b-actin (Primerbank ID 4501885a1). Results are reported as average band integrated intensity +/- SEM. Significance was determined by Student's t-test with p<0.05 considered significant (*).
3. Results
3.1. Characterization of amox-induced persistence in vitro
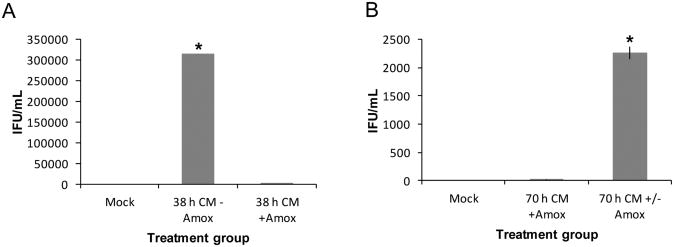
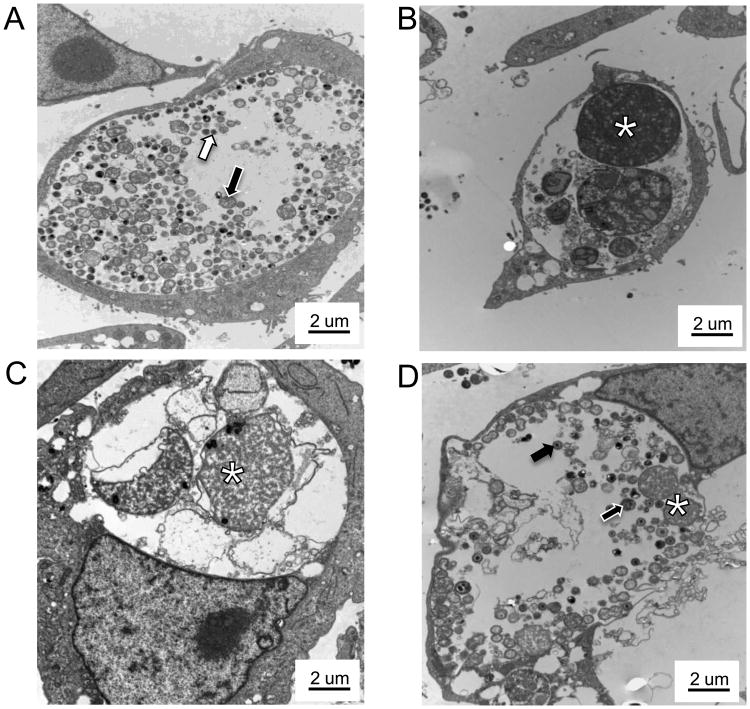
C. muridarum is more than 90% genetically identical to C. trachomatis [27] and produces a similar ascending genital tract disease when inoculated in mice. Amox is known to reduce C. trachomatis titer in culture [28], but infection development following amox exposure in C. muridarum-infected cells is uncharacterized. Our initial experiments, therefore, were designed to test the suitability of amox for inducing persistence in C. muridarum-infected primary genital epithelial cells. BM1.11 monolayers were either mock- or C. muridarum infected, and at 8 hpi, cultures were refed with medium with (+) or without (-) amox. Thirty hours later, replicates were harvested for TEM or titer analyses. Other amox-exposed replicates were refed at 38 hpi with medium either (+) amox (to continue exposure) or (-) amox (to allow recovery) and allowed to incubate until 70 hpi. Unexposed, infected cells at 38 h produced numerous infectious EBs (Fig. 1A) (avg = 314,222 IFU/mL, p<0.05) and contained large inclusions with a mix of RBs, IBs, and EBs (Fig. 2A), as expected during normal development. Mock-infected cells contained no inclusions (Fig. S1). Amox-exposed, C. muridarum-infected cultures at either 38 (Fig. 1A & 2B) or 70 h (Fig. 1B & 2C) contained primarily ABs and produced >99% fewer infectious EBs compared to (-) amox controls harvested at similar time intervals (133 and 11 IFU/mL, respectively). Cells infected with C. muridarum for 30 h (+) amox and allowed to recover in (-) amox medium for 32 h (Fig. 1B & 2D) contained some ABs (Fig. 2D, asterisks) but also both IBs and EBs. As expected, these cells also produced significantly more infectious EBs in titer assays than did infected cells exposed to amox continuously (Fig. 1B) (2,261 IFU/mL versus 11 IFU/mL, p<0.05).
Figure 1.
Amox exposure reversibly reduces C. muridarum infectivity in culture. BM1.11 monolayers were either mock- or C. muridarum (CMW)-infected. At 8 hpi, cultures were refed with medium (+/-) amox; 30 h later, some replicates were harvested for TEM (Fig. 2) or titer analyses (panel A). Other amox-exposed replicates were refed with either antibiotic-free medium (70 h CM +/- Amox) or medium (+) amox (70 h CM + Amox) at 38 hpi and allowed to incubate until 70 hpi (panel B). Triplicate inclusion counts were averaged and used to calculate inclusion forming units (IFU)/mL. The average of three biologic replicates ± SEM from one representative experiment is shown. Groups significantly different from the diluent-exposed, infected control are indicated by asterisks (*), P ≤ 0.05 was considered significant. These results are representative of three independent experiments.
Figure 2.
Amox exposure induces C. muridarum AB formation in culture. Mock or C. muridarum-infected BM1.11 cultures were refed with medium (+/−) amox; 30 h later, some replicates were harvested for TEM while other amox-exposed replicates were refed with either medium or medium (+) amox at 38 hpi and allowed to incubate until 70 hpi. Panel A shows amox (-) infected cells at 38 hpi, panel B shows amox (+) infected cells at 38 hpi, panel C shows amox (+) cells at 70 hpi, and panel D shows cultures exposed to amox from 8 to 38 hpi and then cultured in antibiotic-free medium until 70 hpi. Cell pellets were processed for high contrast TEM as described. Transmission electron micrographs are shown at 7000× magnification; scale bars measure 2 um. RBs are indicated by white-outlined black arrows, EBs by black-outlined white arrows, IBs by solid black arrows and ABs by asterisks. These results are representative of three independent experiments.
3.2. Determination of an in vivo chlamydial inoculum
Eight mice per group were infected with either 104, 105, or 106 IFU of C. muridarum stock diluted in 2SPG. Vaginal swabs were collected every third day and sample dilutions used to infect duplicate coverslips of Hela 229 cells. Inclusions were stained with Pathfinder, counted, and infectious IFU shed per mouse calculated. Peak shedding was obtained on 6 dpi, with no significant differences observed among groups (Fig. S2). An inoculum of 106 IFU per mouse was selected for further studies to increase the probability that inclusions would be found in the cervical or uterine tissue when visualized by TEM.
3.3. Amox treatment induces a non-infectious but viable (persistent) state in vivo
Amoxicillin is an ideal candidate for use in an in vivo persistence model because it: i) is relatively non-toxic, ii) targets developing chlamydiae rather than functioning through host pathways, iii) is not produced naturally in any animal host, iv) is easy to administer, and v) is well understood pharmokinetically. Two mg/kg and 40 mg/kg doses of amox were selected because of our in vitro observations that low doses of amox are adequate to induce persistence in culture. Each of these doses are well below that reported to cause toxicity [29,30]. In the interest of maintaining clinical relevancy, the dosing regimen was chosen to be similar to that used in humans.
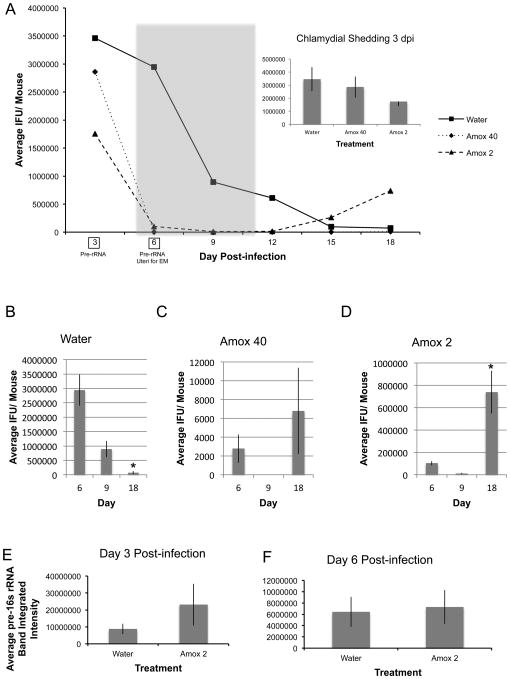
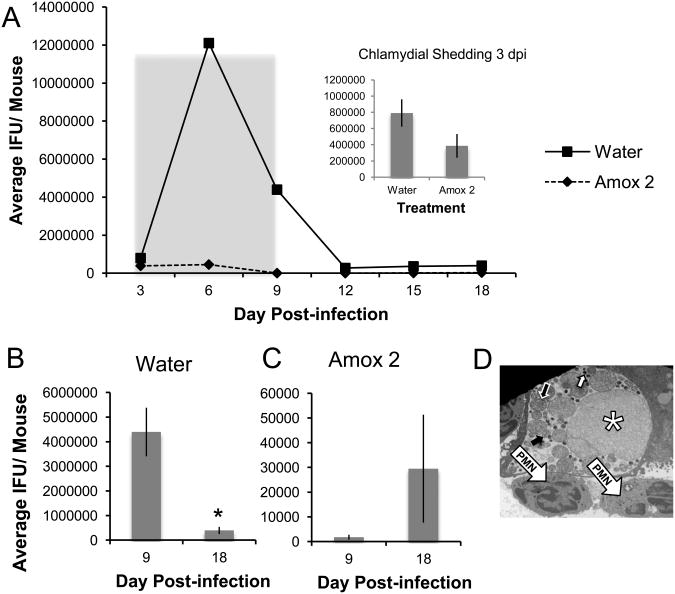
Seven to 12 mice per group were infected with 106 IFU C. muridarum and amox treated beginning at 5 dpi, just before maximum shedding was observed in initial experiments (Fig. S2), and continuing until 11 dpi. Vaginal swabs were collected every third day and titered to determine total infectious EBs shed. Two mice from each group were sacrificed 6 dpi and their uteri processed for TEM. Total RNA and DNA were isolated from day 3 and 6 swab samples (indicated by boxes in Fig. 3A). Chlamydial genomic DNA was quantified by PCR and pre-16S rRNA using RT-PCR as described; pre-16S rRNA was normalized to genomic DNA content as previously reported [25]. Pre-16S rRNA accumulation observed in these experiments is plotted in Fig 3E&F.
Figure 3.
Amox-treatment induces C. muridarum to enter a viable but non-infectious state in the genital tracts of infected mice. Mice were infected with 106 IFU C. muridarum and amox treated from days 5 to 11 pi (panel A, gray shading). Vaginal swabs were collected every third day and titered to determine total infectious EBs shed. The shedding profile over the course of the entire experiment is shown in panel A. The panel A inset as well as panels B through D are comparisons of shedding at day 3 (2 days before treatment), 6 (1 day into the treatment period), 9 (4 days into the treatment period), and day 18 (1 week after treatment termination) for water- (panel B), 40mg/kg amox- (panel C), and 2mg/kg (panel D) amox-gavaged, C. muridarum-infected animals. Triplicate inclusion counts were averaged and used to calculate inclusion forming units (IFU)/mL. The average of 7 mice ± SEM from one representative experiment is shown. Groups significantly different from diluent-exposed, infected control mice are indicated by asterisks (*) in panels B though D, P ≤ 0.05 was considered significant. In panels E and F, RNA and DNA were isolated from day 3 (panel E) and day 6 (panel F) swab samples. Host and chlamydial genomes as well as chlamydial pre-16s ribosomal RNA were quantified as described. Average normalized pre-16S rRNA amplimer intensity from 7 mice per group are shown in panels E and F. The average of 7 mice ± SEM from one representative experiment is shown. Groups significantly different from diluent-exposed, infected control mice are indicated by asterisks (*), P ≤ 0.05 was considered significant. All data are representative of at least three independent mouse infection experiments, each containing 7-10 animals.
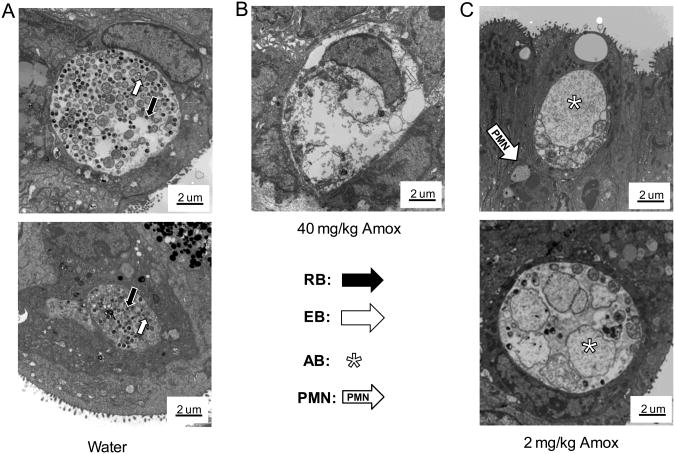
Mock-infected mice, including those receiving amox treatment, did not shed viable organisms (data not shown). No significant differences in infectious shedding were observed among infected groups at the onset of the experiment (Fig. 3A inset). As anticipated, chlamydial shedding decreased over time in mice gavaged with water alone and no secondary peak in shedding was observed at 18dpi (Fig. 3B). Shedding kinetics are similar in control mice that were not gavaged (data not shown). In contrast, amox gavage reduced shedding from infected mice by >90% by day 6 pi (p<0.0001, Fig. 3C&D). Treatment with either amox dose reduced shedding greater than 99% by 9 dpi compared to water gavaged, infected mice (Fig. 3A). Both amox treatments resulted in a secondary peak of shedding by 18 dpi, with 71% of mice in the amox 40 mg/kg group and 100% of mice in the amox 2 mg/kg group resuming shedding (Fig. 3C&D). Inclusions from mice gavaged with water contained normal developmental forms, with a mix of EBs, RBs, and IBs present (Fig. 4A). Mice gavaged with 2 mg/kg amox developed inclusions filled with large, aberrant RBs (Fig. 4C), whereas those gavaged with 40 mg/kg amox had primarily inclusions that appeared to be filled with chlamydial debris (Fig. 4B). Empty inclusions were also observed in uteri from mice treated with the 40 mg/kg dose. Overall, few normal chlamydial forms were observed in amox-treated mice. These data indicate that amox treatment induces formation of aberrant bodies in vivo similar to those observed during persistence in culture (Fig. 2B&C; [6,31]). Electron microscopy and post-treatment shedding recovery data suggest that 2 mg/kg of amox was the optimal “persistence-inducing” dose.
Figure 4.
C. muridarum ABs are present in the genital tracts of amox-treated, infected mice. Two mice from each C. muridarum-infected group were sacrificed 6 dpi (24 h after treatment initiation) and their uteri prefixed and processed for high contrast TEM as described. First, 2 um thick sections were stained with Epoxy Tissue Stain (EM Sciences, Hatfield, PA)[18] and examined by light microscopy at 400× to locate areas in the tissue that might contain inclusions. Areas with visible inclusions were then subjected to additional thin sectioning and TEM. Representative transmission electron micrographs are shown above at 7000× magnification. Scale bars measure 2 um, RBs are indicated by white-outlined black arrows, EBs by black-outlined white arrows, IBs by solid black arrows and ABs by asterisks. The results are representative of three independent experiments.
Pre-16s rRNA is processed co-transcriptionally and, thus, its presence indicates continued transcriptional activity and chlamydial viability [25]. No significant differences in pre-16s rRNA accumulation were observed between groups gavaged with water or amox at either 3 dpi (Fig. 3E, p=0.17) or 6 dpi (Fig. 3F, p=0.42), which represents 24 hours of amox treatment. Pre-16s rRNA accumulation in infected mice receiving water gavage was not significantly different from non-gavaged animals (data not shown, p=0.35 for day 3 and 0.22 for day 6). Because shedding of infectious organisms was reduced by >90% by 2 mg/kg amox treatment at 6 dpi (Fig. 3A), but pre-16s rRNA accumulation at this time was unaltered (Fig. 3E&F), these data suggest that amox treatment induces persistence rather than killing the chlamydiae.
3.4. The persistent state can be induced by an alternate amox treatment schedule
Consistent with previous shedding curves, a strong secondary shedding peak was observed in amox-treated animals after treatment was terminated (Fig. 3A). We reasoned that earlier treatment timing might alter the magnitude of secondary shedding by reducing initial C. muridarum dissemination and/or by altering initial immune activation to the infection. To determine if the day of treatment initiation affected chlamydial shedding and the presence of aberrant forms, 10 mice per group were infected with 106 IFU C. muridarum and amox treated from 3 dpi to 9 dpi. Vaginal swabs were collected every third day and titered as before to determine total infectious EBs shed. Two mice from each group were sacrificed at 6 dpi and uteri processed for TEM. No significant differences in infectious shedding were observed among treatment groups at the onset of the experiment (Fig. 5A inset). As anticipated, chlamydial shedding decreased over time in mice gavaged with water alone, with no secondary peak in shedding (Fig. 5B). Also, no differences were observed between non-gavaged controls and water-gavaged mice (data not shown). Mice that received 2 mg/kg amox gavage shed far fewer infectious chlamydiae (shedding decreased >99%) by 9 dpi compared to the water-gavaged group. As in the previous experiments (Fig. 3A), animals treated with amox at day 3 demonstrated a secondary peak of shedding at 18 dpi (Fig. 5C), with 90% of mice resuming shedding. The secondary shedding peak at 18 dpi was consistently lower in mice that were amox-treated on day 3 (Fig. 5C) compared to that observed when treatment was initiated on day 5 (Fig. 3A). As expected, mice gavaged with amox developed inclusions filled with large, aberrant RBs (Fig. 5D). These results confirm that amox-treatment induces the non-infectious but viable state when amox treatment is initiated at several times post-infection. Furthermore, the magnitude of secondary shedding observed appears to be dependent upon the day post-infection when amox-treatment is initiated.
Figure 5.
Secondary shedding and ABs are also observed in C. muridarum-infected mice when amox treatment is initiated earlier during infection. Mice were infected with 106 IFU C. muridarum and amox treated from day 3 to day 9 pi. Vaginal swabs were collected every third day and titered to determine total infectious EBs shed. The shedding profile over the course of the entire experiment is shown in panel A. The panel A inset as well as panels B and C are comparisons of shedding at day 3 (immediately before the first treatment), day 9 (the last day of treatment) and day 18 (9 days after treatment termination) for water- (panel B) and 2mg/kb amox-gavaged (panel C) C. muridarum-infected animals. Triplicate inclusion counts were averaged and used to calculate inclusion forming units (IFU)/mL. The average of 10 mice ± SEM from one of two independent experiments is shown. Groups significantly different from diluent-exposed, infected control mice are indicated by asterisks (*) in panels B and C, P ≤ 0.05 was considered significant. Panel D shows a representative TEM photomicrograph of genital tract tissue isolated from a 2mg/kg treated, C. muridarum-infected mouse at 5 dpi (2 days after treatment initiation). Transmission electron micrographs are shown at 7000× magnification; scale bars measure 2 um. RBs are indicated by white-outlined black arrows, EBs by black-outlined white arrows, IBs by solid black arrows, and ABs by asterisks. White arrow indicates PMN.
4. Discussion
Previous studies have suggested that viable, but non-infectious chlamydiae are present during in vivo infection. Continued and recurrent infections with the same serovar occur in patients who have completed the recommended treatment regimens [7–10], viable chlamydiae are observed in patients with C. trachomatis-induced reactive arthritis [14], and chlamydiae showing morphologic characteristics of persistent organisms are present in tissue samples [15–18]. Additional evidence includes recurring clinical signs of infection in infected cats and mice treated with clavulanate-potentiated amoxicillin [19,20] and the presence of chlamydial DNA, RNA, and antigens in mice and non-human primates subsequent to culture-apparent resolution [11–13]. Though the conclusions of each of these studies are consistent with persistence induction in vivo, the investigators' use of either TEM or pre-16S rRNA analyses, but not both, incompletely defines persistence.
In these experiments, our laboratory has demonstrated that amoxicillin administration significantly reduced the number of infectious chlamydiae shed while chlamydial pre-16s rRNA accumulation was unaffected, demonstrating that chlamydiae that are amox-exposed in vivo are viable but non-infectious. Further, mice gavaged with water do not resume shedding even when followed out to 55 dpi (data not shown), reducing the possibility that levels of normally differentiating EBs below our detection levels are responsible for the secondary peak observed in amox-gavaged mice. Additionally, all inclusions observed by TEM in the genital tracts of 2 mg/kg amox-treated animals contain primarily aberrant chlamydial forms (ABs) and none contained EBs. These results are identical to those obtained when chlamydia-infected cells are exposed to amox in culture (Fig. 2), as well as to data obtained from other culture models of persistence (2). Taken together, these data definitively demonstrate that persistence occurs in vivo in response to amoxicillin. Additionally, these data indicate that amox treatment induces persistence under several different dose and timing regimens. Finally, we commonly observed PMNs in close proximity to AB-containing inclusions in amox-treated, chlamydial infected genital tract samples (Fig. 4C&5D), which is consistent with the prediction by Beatty et al. that the presence of chlamydial antigens in the absence of cultivatable organisms may continue to result in immunologic stimulation and disease [21].
Not surprisingly, the magnitude of post-treatment secondary shedding appears to be dependent upon treatment timing. Treatment at day 5 likely allows greater dissemination of the chlamydiae compared to that in animals on a day 3 treatment schedule. This would increase the number of infected genital cells present when treatment is initiated and likely increase the size of the secondary shedding peak. Early treatment may also limit or alter initial immune activation, which could, in turn, alter post-treatment secondary shedding kinetics. It is also important to note that in animals treated at day 5 pi, the secondary shedding peak never reached pre-treatment shedding levels. Development of adaptive immunity over the course of the experiment may, in part, explain this observation. However, this smaller secondary peak (Fig. 3A) is consistent with the in vitro recovery data (Fig 1B), which suggest that recovery from amox-induced persistence is incomplete even in the absence of host immunity. It seems likely that a significant proportion of amox-exposed chlamydial ABs are “locked” in the non-replicative state and cannot continue normal development even upon drug removal. This prediction is supported by published in vitro observations that not all inclusions resume the developmental cycle when penicillin is removed [31], as well as by our data suggesting that lower doses of amox allow for increased secondary shedding of infectious chlamydiae post-treatment than do higher doses (Fig. 3 C&D, Fig. S3). Ongoing studies in our laboratory seek to determine how each of these factors contribute to chlamydial recovery from the persistent state in vivo.
Chlamydial persistence induction in culture by stimuli such as nutrient deprivation, IFN-gamma, and penicillins is well defined. Importantly, persistent organisms in culture are more resistant to killing by anti-chlamydial drugs [32,33] than are non-persistent forms. Persistent chlamydiae also inhibit host cell apoptosis, demonstrating that persistent organisms can alter host cellular processes [34,35]. Persistence also increases production of pro-inflammatory molecules, such as chlamydial HSP60 (cHSP60) [2]. While not definitive, in vitro data suggest that persistent organisms may contribute to the long-term inflammation, fibrosis, and scarring characteristic of chlamydial diseases by serving as a reservoir for chlamydial antigens, pro-inflammatory effectors, and continued productive infection, as well as inducing sustained host cell physiologic dysfunction. Though it may require additional optimization, our in vivo model provides the first system in which the role of aberrant forms in disease production, as well as in establishment of continuing or recurring infections, can be addressed.
This murine model of chlamydial persistence not only allows study of the effects of persistence in vivo, but also potentially has significant clinical implications. As it is the most commonly prescribed beta-lactam in the U.S., amox may be given unknowingly to a patient with asymptomatic infection, and it is still a medication of choice for the treatment of chlamydial infection in pregnant patients [36]. Its ability to induce persistence in mice, particularly in regard to post-treatment secondary shedding, underlines the importance of screening and follow-up testing when amox is used as primary treatment for chlamydial genital infection. Notably, current CDC recommendations call for repeat testing, preferably by NAAT, of pregnant women to document chlamydial eradication 3 weeks after completion of therapy [37].
Supplementary Material
Supplemental Figure 1. TEM of mock-infected BM1.11 cells.
Supplemental Figure 2. Course of infection for multiple C. murdarum inocula. Eight mice per group were infected with either 104, 105, or 106 IFU of C. muridarum stock diluted in 2SPG. Vaginal swabs were collected every third day and dilutions used to infect Hela 229 cells. Inclusions were fluorescently stained, counted, and multiplied by dilution and collection volume to obtain IFU per mouse. Two coverslips per mouse were averaged.
Supplemental Figure 3. Secondary shedding is also observed in C. muridarum-infected mice with lower dose amox treatment. Mice were infected with 106 IFU C. muridarum and amox treated with either 2 mg/kg or 02. mg/kg from day 5 to day 11 pi. Vaginal swabs were collected every third day and titered to determine total infectious EBs shed.
Acknowledgments
The authors want to thank Dr. Priscilla B. Wyrick (East Tennessee State University; ETSU), the ETSU Animal Care Facility staff, Dr. Kyle Ramsey (Chicago College of Osteopathic Medicine, Midwestern University), and Dr. Ray Johnson (Indiana University School of Medicine) for providing reagents and for their many helpful discussions. We also want to acknowledge the Electron Microscopy Core Facility (ETSU, Quillen College of Medicine). This work was supported by NIH/NIAID R15 #AI078373-01A1 and NIH/NIAID R21 #AI082322-01 to R.V.S. and an ETSU. School of Graduate Studies Research Award to R.B.P.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schoborg RV. Chlamydia persistence -- a tool to dissect chlamydia--host interactions. Microbes Infect. 2011;13:649–662. doi: 10.1016/j.micinf.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco JA, Tan C, Rank RG, Hsia RC, Bavoil PM. Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis. Cell Microbiol. 2011;13:1014–1025. doi: 10.1111/j.1462-5822.2011.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatty WL, Morrison RP, Byrne GI. Reactivation of Persistent Chlamydia trachomatis infection in cell culture. Microbiol. 1995;63:199–205. doi: 10.1128/iai.63.1.199-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galasso GJ, Manire GP. Effect of antiserum and antibiotics on persistent infection of HeLa cells with meningopneumonitis virus. J Immunol. 1961;86:382–385. [PubMed] [Google Scholar]
- 6.Matsumoto A, Manire GP. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol. 1970;101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortenberry JD, Brizendine EJ, Katz BP, Wools KK, Blythe MJ, Orr DP. Subsequent sexually transmitted infections among adolescent women with genital infection due to Chlamydia trachomatis, Neisseria gonorrhoeae, or Trichomonas vaginalis. Sex Transm Dis. 1999;26:26–32. doi: 10.1097/00007435-199901000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Patton DL, Askienazy-Elbhar M, Henry-Suchet J, Campbell LA, Cappuccio A, Tannous W, Wang SP, Kuo CC. Detection of Chlamydia trachomatis in fallopian tube tissue in women with postinfectious tubal infertility. Am J Obstet Gynecol. 1994;171:95–101. doi: 10.1016/s0002-9378(94)70084-2. [DOI] [PubMed] [Google Scholar]
- 9.Dean D, Suchland RJ. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. Infect Immun. 2000;182:909–916. doi: 10.1086/315778. [DOI] [PubMed] [Google Scholar]
- 10.Stamm WE. Chlamydia trachomatis--the persistent pathogen: Thomas Parran Award Lecture. Sex Transm Dis. 2001;28:684–689. doi: 10.1097/00007435-200112000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Holland SM, Hudson AP, Bobo L, Whittum-hudson JA, Viscidi RP, Quinn TC, Taylort HR. Demonstration of Chlamydial RNA and DNA during a culture-negative state. Infection. 1992;60:2040–2047. doi: 10.1128/iai.60.5.2040-2047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsey KH, Miranpuri GS, Sigar IM, Ouellette S, Byrne GI. Chlamydia trachomatis persistence in the female mouse genital tract: inducible nitric oxide synthase and infection outcome. Infect Immun. 2001;69:5131–5137. doi: 10.1128/IAI.69.8.5131-5137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bin XX, Wolf K, Schaffner T, Malinverni R. Effect of azithromycin plus rifampin versus amoxicillin alone on eradication and inflammation in the chronic course of Chlamydia pneumoniae pneumonitis in mice. Antimicrob Agents Chemother. 2000;44:1761–1764. doi: 10.1128/aac.44.6.1761-1764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gérard HC, Branigan PJ, Schumacher HR, Hudson AP. Synovial Chlamydia trachomatis in patients with reactive arthritis/Reiter's syndrome are viable but show aberrant gene expression. J Rheumatol. 1998;25:734–742. [PubMed] [Google Scholar]
- 15.Nanagara R, Li F, Beutler A, Hudson HR, Schumacher HR. Alteration of Chlamydia trachomatis biologic behavior in synovial membranes. Suppression of surface antigen production in reactive arthritis and Reiter's syndrome. Arthritis Rheum. 1995;38:1410–1417. doi: 10.1002/art.1780381008. [DOI] [PubMed] [Google Scholar]
- 16.Skowasch D, Yeghiazaryan K, Schrempf S, Golubnitschaja O, Welsch U, Preusse CJ, Likungu JA, Welz A, Lüderitz B, Bauriedel G. Persistence of Chlamydia pneumoniae in degenerative aortic valve stenosis indicated by heat shock protein 60 homologues. J Heart Valve Dis. 2003;12:68–75. [PubMed] [Google Scholar]
- 17.Pospischil A, Borel N, Chowdhury EH, Guscetti F. Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet Microbiol. 2009;135:147–156. doi: 10.1016/j.vetmic.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 18.Rank RG, Whittimore J, Bowlin AK, Wyrick PB. In vivo ultrastructural analysis of the intimate relationship between polymorphonuclear leukocytes and the chlamydial developmental cycle. Infect Immun. 2011;79:3291–3301. doi: 10.1128/IAI.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturgess CP, Gruffydd-Jones TJ, Harbour DA, Jones RL. Controlled study of the efficacy of clavulanic acid-potentiated amoxycillin in the treatment of Chlamydia psittaci in cats. Vet Rec. 2001;149:73–76. doi: 10.1136/vr.149.3.73. [DOI] [PubMed] [Google Scholar]
- 20.Beale AS, Upshon PA. Characteristics of murine model of genital infection with Chlamydia trachomatis and effects of therapy with tetracyclines, amoxicillin-clavulanic acid, or azithromycin. Antimicrob Agents Chemother. 1994;38:1937–1943. doi: 10.1128/aac.38.9.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyrick PB. Chlamydia trachomatis persistence in vitro: an overview. J infect Dis. 2010;201 Suppl:S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guseva NV, Dessus-Babus S, Moore CG, Whittimore JD, Wyrick PB. Differences in Chlamydia trachomatis serovar E growth rate in polarized endometrial and endocervical epithelial cells grown in three-dimensional culture. Infect Immun. 2007;75:553–564. doi: 10.1128/IAI.01517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson RM. Murine Oviduct Epithelial Cell Cytokine Responses to Chlamydia muridarum Infection Include Interleukin-12 – p70 Secretion. Infect Immun. 2004;72:3951–3960. doi: 10.1128/IAI.72.7.3951-3960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deka S, Vanover J, Dessus-Babus S, Whittimore J, Howett MK, Wyrick PB, Schoborg RV. Chlamydia trachomatis enters a viable but non-cultivable (persistent) state within herpes simplex virus type 2 (HSV-2) co-infected host cells. Cell Microbiol. 2006;8:149–162. doi: 10.1111/j.1462-5822.2005.00608.x. [DOI] [PubMed] [Google Scholar]
- 26.Gérard HC, Whittum-Hudson JA, Hudson AP. Genes required for assembly and function of the protein synthetic system in Chlamydia trachomatis are expressed early in elementary to reticulate body transformation. Mol Genet Genomics. 1997;255:637–642. doi: 10.1007/s004380050538. [DOI] [PubMed] [Google Scholar]
- 27.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, Deboy R, Kolonay J, Mcclarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowie WR. In vitro activity of clavulanic acid, amoxicillin, and ticarcillin against Chlamydia trachomatis. Antimicrob Agents and Chemother. 1986;29:713–715. doi: 10.1128/aac.29.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comber KR, Osborne CD, Sutherland R. Comparative effects of amoxycillin and ampicillin in the treatment of experimental mouse infections, Antimicrob. Agents and Chemother. 1975;7:179–185. doi: 10.1128/aac.7.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J Gisby, Beale aS. Comparative efficacies of amoxicillin-clavulanic acid and ampicillin-sulbactam against experimental Bacteroides fragilis-Escherichia coli mixed infections. Antimicrob Agents and Chemother. 1988;32:1830–1833. doi: 10.1128/aac.32.12.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skilton RJ, Cutcliffen LT, Barlow D, Wang Y, Salim O, Lambden PR, Clarke IN. Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS ONE. 2009;4:e7723. doi: 10.1371/journal.pone.0007723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reveneau N, Crane DD, Fischer E, Caldwell HD. Bactericidal activity of first-choice antibiotics against gamma interferon-induced persistent infection of human epithelial cells by Chlamydia trachomatis. Antimicrob Agents Chemother. 2005;49:1787–1793. doi: 10.1128/AAC.49.5.1787-1793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyrick PB, Knight ST. Pre-exposure of infected human endometrial epithelial cells to penicillin in vitro renders Chlamydia trachomatis refractory to azithromycin. J Antimicrob Chemother. 2004;54:79–85. doi: 10.1093/jac/dkh283. [DOI] [PubMed] [Google Scholar]
- 34.Dean D, Powers VC. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect Immun. 2001;69:2442–2447. doi: 10.1128/IAI.69.4.2442-2447.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perfettini JL, Darville T, Dautry-Varsat A, Rank RG, Ojcius DM. Inhibition of apoptosis by gamma interferon in cells and mice infected with Chlamydia muridarum (the mouse pneumonitis strain of Chlamydia trachomatis) Infect Immun. 2002;70:2559–2565. doi: 10.1128/IAI.70.5.2559-2565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anon Chlamydia trachomatis and pregnancy. Prescrire Int. 2011;20:302. [PubMed] [Google Scholar]
- 37.Workowski KA, Berman S. Centers for Disease Control and Prevention (CDC) MMWR Recomm Rep. 2010;59:1–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. TEM of mock-infected BM1.11 cells.
Supplemental Figure 2. Course of infection for multiple C. murdarum inocula. Eight mice per group were infected with either 104, 105, or 106 IFU of C. muridarum stock diluted in 2SPG. Vaginal swabs were collected every third day and dilutions used to infect Hela 229 cells. Inclusions were fluorescently stained, counted, and multiplied by dilution and collection volume to obtain IFU per mouse. Two coverslips per mouse were averaged.
Supplemental Figure 3. Secondary shedding is also observed in C. muridarum-infected mice with lower dose amox treatment. Mice were infected with 106 IFU C. muridarum and amox treated with either 2 mg/kg or 02. mg/kg from day 5 to day 11 pi. Vaginal swabs were collected every third day and titered to determine total infectious EBs shed.