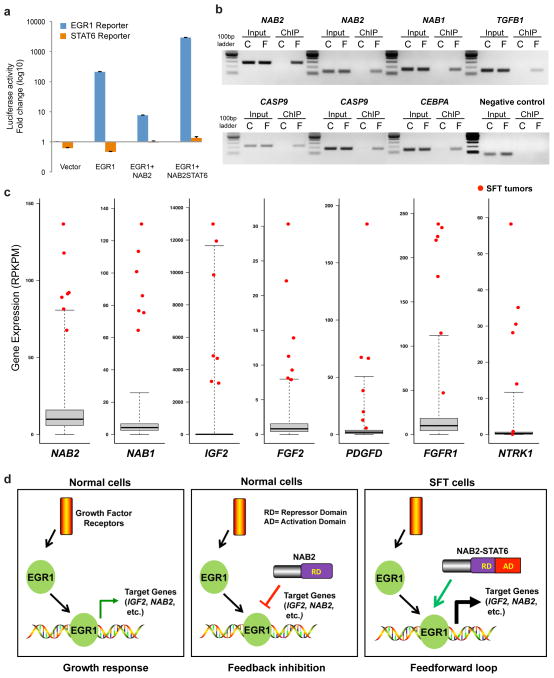
Figure 4.
A proposed model for the function of the NAB2-STAT6 gene fusion in SFT. (a) NAB2-STAT6 fusion protein enhances EGR1-induced promoter activity. We transfected 293T cells with vectors expressing the indicated proteins along with a constitutively expressed Renilla luciferase construct and one of the two reporter constructs EGR1-responsive firefly luciferase reporter or STAT6-responsive firefly luciferase reporter. Cell lysates were prepared 24 h after transfection and measured for EGR1 and STAT6 activity. Means of quadruplicates with the s.d. are shown. (b) ChIP identified the recruitment of NAB2-STAT6 fusion protein to the promoters of EGR1-responsive genes. We transfected 293T cells with only vector expressing EGR1 (C) or cotransfected with vectors expressing EGR1 and Flag-NAB2-STAT6 (F). Crosslinked DNA-protein lysates were immunoprecipitated with antibody to Flag. The promoter fragments containing consensus EGR1-binding sites were PCR amplified from both samples. Input samples were loaded as a control. IGX1A primers that detect a specific genomic DNA sequence within an ORF-free intergenic region were used as a negative control. Two distinct EGR1-binding sites in NAB2 and CASP9 are shown. (c) Outlier gene expression in SFT predicted to be the result of NAB2-STAT6–driven constitutive activation of EGR1-mediated pathways. Data from RNA-seq analysis of 7 SFTs relative to a compendium of 282 previously sequenced tumor tissues are shown (Supplementary Table 6). RPKPM (reads per kilobase per million reads) values for the compendium are summarized by box-and-whisker plots showing the minimum, quartile 1, median, quartile 3 and maximum observed expression levels across the compendium. RPKPM values for the seven SFT samples are shown in red. (d) Model of the NAB2 and EGR1 signaling loops. RD, repressor domain; AD, activation domain.