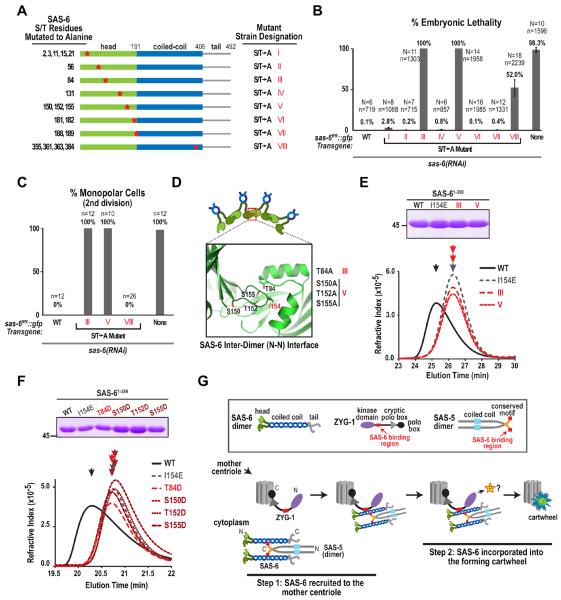
Figure 7. A serine/threonine mutational scan of SAS-6 and a model for cartwheel assembly.
(A) Serines and threonines conserved in Caenorhabditis species (elegans, briggsae, brenneri, japonica, remanei; Fig. S7A) were mutated as indicated in the schematics, to generate 8 transgenic SAS-6∷GFP strains. (B & C) Plots of percent embryonic lethality (B) and frequency of monopolar second division (C), as in Fig. 1H, for the indicated conditions. (D) Schematic showing the location of serine/threonine residues in the crystal structure of the SAS-6 N-terminus (PDB=3PYI) that when mutated to alanine in strains III and V resulted in 100% embryonic lethality and 100% monopolar second divisions. (E & F) Analysis of SAS-6 inter-dimer interactions by gel filtration chromatography. (top panels) Coomassie-stained gels showing the purified SAS-61–389 variants tested in each experiment (see also Fig S7B). (bottom panels) Elution profiles for the purified proteins on a Superdex 200 (E) or a WTC-030S5 (F) size exclusion column. (G) Model for cartwheel assembly in vivo. A direct interaction between ZYG-1 and the SAS-6 coiled-coil recruits the SAS-6—SAS-5 complex to the mother centriole where ZYG-1 kinase activity promotes cartwheel assembly by phosphorylating a target that is unlikely to be SAS-6. See also Figure S7.