Abstract
Background
The Epilepsy Phenome/Genome Project (EPGP) is a large-scale, multi-institutional, collaborative network of 27 epilepsy centers throughout the U.S., Australia, and Argentina, with the objective of collecting detailed phenotypic and genetic data on a large number of epilepsy participants. The goals of EPGP are (1) to perform detailed phenotyping on 3750 participants with specific forms of non-acquired epilepsy and 1500 parents without epilepsy, (2) to obtain DNA samples on these individuals, and (3) to ultimately genotype the samples in order to discover novel genes that cause epilepsy. To carry out the project, a reliable and robust informatics platform was needed for standardized electronic data collection and storage, data quality review, and phenotypic analysis involving cases from multiple sites.
Methods
EPGP developed its own suite of web-based informatics applications for participant tracking, electronic data collection (using electronic case report forms/surveys), data management, phenotypic data review and validation, specimen tracking, electroencephalograph and neuroimaging storage, and issue tracking. We implemented procedures to train and support end-users at each clinical site.
Results
Thus far, 3780 study participants have been enrolled and 20,957 web-based study activities have been completed using this informatics platform. Over 95% of respondents to an end-user satisfaction survey felt that the informatics platform was successful almost always or most of the time.
Conclusions
The EPGP informatics platform has successfully and effectively allowed study management and efficient and reliable collection of phenotypic data. Our novel informatics platform met the requirements of a large, multicenter research project. The platform has had a high level of end-user acceptance by principal investigators and study coordinators, and can serve as a model for new tools to support future large scale, collaborative research projects collecting extensive phenotypic data.
Keywords: EPGP, Informatics, Epilepsy, Research, Internet, Electronic data collection, Phenotyping
1. Introduction
1.1. Background
The Epilepsy Phenome/Genome Project (EPGP) [1] is a large-scale, multi-institutional, collaborative network consisting of 27 epilepsy centers throughout the U.S., Australia, and Argentina, with the objective of collecting detailed phenotypic and genetic data on a large number of epilepsy participants. The goals of EPGP are (1) to perform detailed phenotyping on 3750 participants with specific forms of non-acquired epilepsy and 1500 parents without epilepsy, (2) to obtain DNA samples on these individuals, and (3) to ultimately genotype the samples in order to discover novel genes that cause epilepsy. Specifically, EPGP will characterize the clinical, electrophysiological and neuroimaging phenotypes of participants with discrete subtypes of idiopathic generalized, localization-related or severe early-onset epilepsy, and use these data in combination with genomic analyses to identify genetic determinants of these phenotypes.
Because EPGP is collecting large amounts of phenotypic, imaging and genomic data on thousands of study participants, a reliable and robust informatics platform was deemed essential to maximize efficiency [2] and security of data collection. An informal assessment of what technologies were used on other clinical research projects found that commercial solutions were too expensive and open-source solutions did not adequately provide the functionality required to support the needs of the EPGP study. Therefore, we created sophisticated web-based applications for participant tracking, electronic data collection (using electronic case report forms/surveys), data management, phenotypic data review and validation, specimen tracking, and electroencephalography (EEG) and magnetic resonance imaging (MRI) storage and review. A central EPGP data repository was designed to store all transactional data, and a data warehouse was created to facilitate reporting, data exploration, and data analysis. Procedures were also developed to train and support end-users at each clinical site. Participant privacy was paramount, so EPGP’s systems met all the requirements of current privacy standards, such as the HIPAA (Health Insurance Portability and Accountability Act) [3] and 21CFR Part 11 [4].
This report outlines the applications and tools we developed for EPGP, the support structures created, the results of the informatics platform implementation, results of an end-user satisfaction survey, lessons learned, and plans for future development.
2. Methods
2.1. Protocol overview
Participants meeting study eligibility criteria [5] are interviewed, and two tubes of blood are drawn and sent to the NINDS Human Genetics Resource Center DNA and Cell Line Repository at the Coriell Institute for Medical Research [6] in Camden, NJ for DNA extraction and storage. Detailed information is collected on demographic variables, the history of seizure disorders and relevant medical history, and medication response, using standardized interviews, medical record abstraction, and collection and review of primary data from EEGs and MRIs. All of this information is reviewed by a site principal investigator (PI) to arrive at a classification of seizure type and epilepsy syndrome, which can be reviewed again at a later stage by the Data Review Core for accuracy. Participants who are unable to come to an EPGP clinical center can have the blood draw performed by a phlebotomy service company at the participants’ homes.
2.2. End-users
The primary users of the EPGP informatics platform are PIs and study coordinators. The duties of the study coordinators include enrolling new study participants, conducting web-based surveys and uploading EEG files, MRI files and medical records. PIs and Co-PIs complete the Final Diagnosis Forms for their site’s study participants, and complete the characterization of their participants’ EEGs and MRIs.
2.3. System development
After searching for suitable off-the-shelf and open-source tools to support the informatics needs of EPGP, we decided to develop our own suite of informatics applications based on the Microsoft® platform. This approach allowed for the most flexibility in terms of ease of use, and availability of training materials to train end-users, which most third-party systems do not provide [7]. Because of a limited timeframe of 12 months for the development of all applications, we adopted a rolling-wave approach to creating the software that involved a progressive elaboration of system development. This enabled us to deploy the applications on a staggered basis. The software design process used physical prototypes of the web applications rather than relying on verbose design specifications, and this permitted us to develop the software more quickly and respond to changing requirements in an agile manner. There is a single central EPGP data repository that stores all the study’s phenotypic data (transactional data), but because of its complexity, it did not lend itself easily to interface with external reporting and data analysis tools. Therefore, we designed and implemented a separate EPGP data warehouse that is populated from the transactional database using ETL (Extract, Transform, Load) techniques on a near real-time basis. The EPGP data warehouse is fully documented in a data dictionary that explains all of the data elements, ensuring that the data warehouse is easily comprehensible to the researchers who use it.
2.4. Application architecture
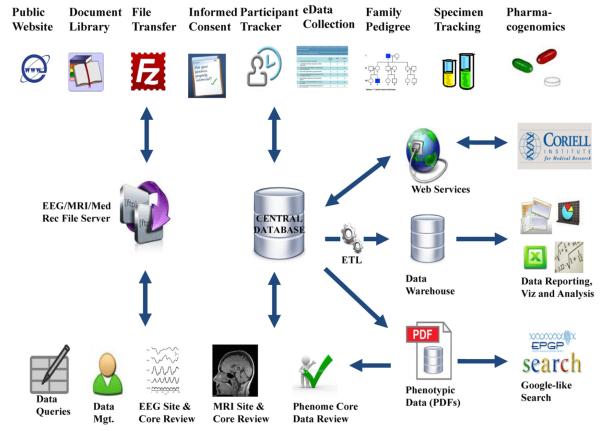
Fig. 1 depicts the suite of web-based applications that comprise the EPGP informatics platform, along with the interaction between the applications and the databases illustrating data flow and functionality.
Fig. 1.
EPGP application architecture.
2.5. Document library/team collaboration
The EPGP project uses Microsoft® SharePoint® 2010 [8] for team collaboration, file sharing and document management. Features implemented include an EPGP project collaboration website, discussion boards, contact lists and news items. The document management feature is used to store the study’s essential documents, such as the study protocol, manual of operations (MOP), standard operating procedures (SOPs), training videos and meeting minutes.
2.6. Participant Activity Tracker
The Participant Activity Tracker (PAT) application allows study personnel to enroll new participants, record phenotypic data and track the progress of the participants’ phenotyping activities, such as data collection forms, blood specimens, AED (antiepileptic drug) histories, EEG reviews and MRI reviews. Electronic data collection forms are launched in a web browser and are completed entirely online with the participant present (or remotely via a telephone interview). The study coordinator can search for their site’s participants’ records using different search criteria and update the participant’s information (such as epilepsy type, participant type, age, gender and initials), as well as generate family pedigree charts. The subject’s participant type (e.g. proband, sibling, affected parent, unaffected parent) determines which versions of the questionnaires are used. The last activity for each participant is the Final Diagnosis Form, which requires all other activities to be completed first.
2.7. Specimen tracking
Blood specimens are collected on all study participants and are stored at Coriell. Each specimen collection tube is tagged with the participant’s identifier and a unique tube identifier, which is used to track the specimen throughout its lifecycle. The submitting clinical site monitors the specimen shipment using EPGP’s web-based specimen tracking system, which has an interface to the courier’s web service to track each shipment during transit. When the specimen is received at Coriell, their IT system calls EPGP’s web service to record that the specimen was received and passes back the Coriell/NINDS identifier assigned to the participant’s specimen.
2.8. Family pedigree
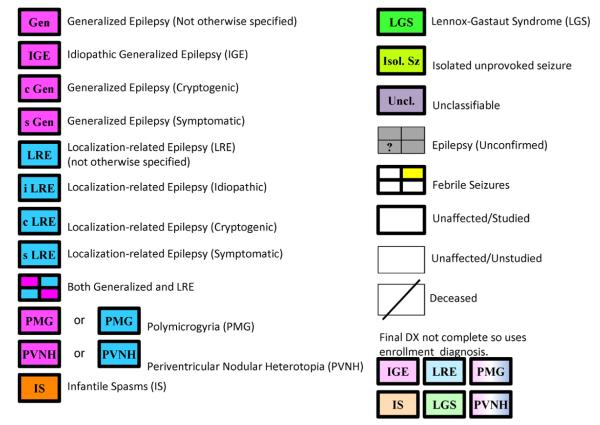
The family pedigree chart is a pictorial display of a participant’s family relationships and epilepsy history. EPGP’s pedigree charting tool was developed using Microsoft® Silverlight® [9], and is integrated into the Participant Activity Tracker (PAT) web application. It allows family members enrolled in the study to be linked using an easy-to-use interface, and accommodates families consisting of multiple generations and extended family members. As shown in Fig. 2, the symbols used to represent participants in the family pedigree are drawn using heavy lines to indicate the individual is an enrolled participant. Bold or dark color-fills indicate that the participant’s epilepsy type has been taken from the participant’s Final Diagnostic Form (completed after all data have been collected and reviewed), which also contains the participant’s International League Against Epilepsy (ILAE) code [10] for epilepsy syndrome classification. The lighter/faded color-fills indicate that the participant’s Final Diagnosis Form has not yet been completed, and the epilepsy type was obtained from the data collected at enrollment instead (Fig. 3).
Fig. 2.
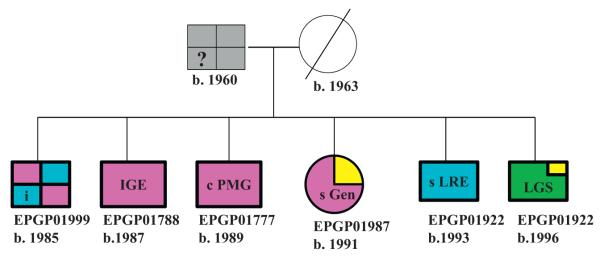
Example pedigree chart.
Fig. 3.
EPGP pedigree chart symbols.
2.9. Web-based phenotypic data collection
EPGP’s web-based platform enables full tracking of the study’s participants across multiple clinical sites and provides for 100% electronic data collection (EDC). Electronic Case Report Forms (eCRFs) are designed using a web-based forms designer, which can accommodate a broad range of question types (e.g. free text, radio buttons, checkboxes, drop-down lists and grid questions). It has advanced data validation and branching capabilities to streamline the data collection process and ensure that data are collected correctly.
The eCRFs are launched from within the Participant Activity Tracker application by clicking on a specific activity, as shown in Fig. 4. The list of activities is specific to the selected participant (i.e. the participant’s schedule of activities is based on their participant type and epilepsy type). The completion status of an activity can be ‘Not started’, ‘In-progress’, ‘Site completed’ and ‘Completed’. If the status of the eCRF activity is ‘In-progress’, the end-user can resume the form from the point he/she left off. The status of the CRF will automatically change to ‘Completed’ when the activity is entirely finished. An activity that is ‘Site-completed’ means that the clinical site has completed the activity, but it has been forwarded to a core team for further review (e.g. EEG or MRI Core Review).
Fig. 4.
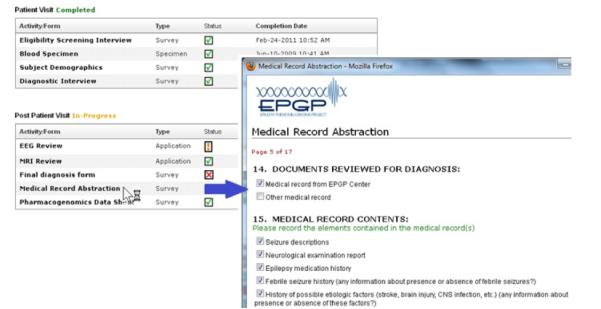
Launching an electronic case report form.
2.10. Data warehouse
The EPGP data warehouse was implemented using Microsoft® SQL 2008® [11] and is used primarily to facilitate intuitive reporting, analysis and data visualization. Essential components of the data warehouse architecture are the ETL (Extract, Transform, Load) processes that populate the data warehouse regularly, and the data dictionary that describes the data elements in the data warehouse. The ETL processes are automated and import phenotypic data from the transactional databases every four hours so that the data warehouse is near real-time. The EPGP data dictionary is a catalogue of all EPGP’s clinical data elements in the data warehouse. It consists of metadata that describes the underlying data, and is essential because of the volume and diversity of data stored in the data warehouse. The data dictionary consists of more than 3000 clinical data elements and contains information such as data point name, data type, length, description, origin, usage, format and encoding.
2.11. EEG/MRI file store
Clinical sites submit primary EEG and MRI files electronically over a secure channel to EPGP’s FTP file server, or via a courier or mail. The file transfer protocol uses FTPS (also known as FTP Secure and FTP-SSL), which is an extension to the commonly used file transfer protocol (FTP). FTPS adds support for the Transport Layer Security (TLS) and the Secure Sockets Layer (SSL) to provide secure communications over the Internet. Only authenticated end-users using a client FTP application that supports FTPS-IMPLICIT can connect to EPGP’s FTP file server.
2.12. EEG/MRI phenotyping and core review
Participants’ EEG and MRI studies are collected to confirm epilepsy diagnosis and classification, and to facilitate genetic analysis of specific EEG and MRI features. The EEG and MRI files are uploaded to a ‘drop zone’ on the FTP file server. From here, the files are de-identified and archived into a standard accessible format. EEGs are archived to Persyst Insight II® [12] format and MRIs are stored in DICOM (Digital Imaging and Communications in Medicine) [13] format. When the files are ready for characterization by the respective EEG or MRI Review Cores, a workflow is triggered and the randomly selected reviewers are alerted via email. The EEG/MRI reviewers use the appropriate software to view the EEG or MRI, and use EPGP’s web-based review applications to record the phenotypic characteristics. Where there are disagreements on the EEG/MRI phenotyping or the tests are deemed ineligible for inclusion in the study, a Consensus Review process takes place, allowing a final decision to be taken on the specific EEG/MRI. The ability to use web-based tools to capture and review EEG/MRI phenotypic data ensures that the accuracy of phenotyping and eligibility standards are upheld, and it promotes consistent EEG/MRI phenotyping for all study participants [14].
2.13. Pharmacogenomics
We developed novel informatics tools for the recording and reviewing of responses to AEDs. These tools facilitate the rigorous collection of drug response data, and careful assignment of pharmacosensitive or pharmacoresistant phenotypes of a large sample of participants with epilepsy. These data will enable the detection of clinically meaningful associations between multi-gene mutations and AED resistance. The tools include (1) a decision tree to classify the AED, which leads the study coordinator through a series of decision points that will ultimately lead to the AED classification of uninformative, success or failure, (2) a web-based data collection form to collect the participant’s AED medication and drug response history, and (3) a web-based AED Review process to allow the AED Core review all AED data.
2.14. Phenotypic data review
The purpose of the phenotypic data review application is to facilitate a review of medical records and phenotypic data for a subset of EPGP’s study participants. As depicted in the workflow shown in Fig. 5, two initial reviewers are randomly selected to review each participant’s data. If either of these reviewers flags any concerns in the data, an automatic email, which contains details of the concern(s) recorded, is sent to the clinical site PI. Through an exchange of emails between the clinical site PI and the primary data reviewer, the reviewer eventually decides what changes (if any) are required to the participant’s data; these are then implemented by the clinical site PI by editing the existing data using the web-based phenotypic forms.
Fig. 5.
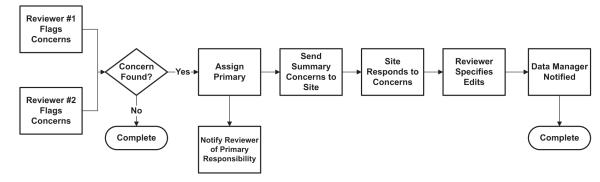
Data review workflow.
2.15. Data reporting/data visualization
The phenotypic data in the database are accessible through a variety of means, including a ‘Google-like’ search web site, MS Excel/MS Access and various other data analysis and data visualization tools. The ‘Google-like’ search website allows the user to search and retrieve participants’ phenotypic data, which are stored in readable PDF files. End-user database and analysis tools, such as MS Excel/MS Access, can connect directly to the EPGP database via ODBC (Open Database Connectivity), which is a standard software interface for accessing database management systems, over a secure Virtual Private Network (VPN) connection. Access to actual EEGs and MRIs is via a secure FTP client or a mapped-drive to EPGP’s FTP file server.
The data visualization tools enable data stored in EPGP’s large phenotypic datasets to be condensed into meaningful visual representations and facilitate visual comparisons of data. A number of reports and charts were developed so that the study’s progress could be monitored in real-time. Examples of some of the reports that were created are listed in Table 1.
Table 1.
EPGP reports.
| EPGP reports |
| Activities completion by site |
| Adjudications report |
| Billable activities by site |
| Completed participants |
| Completed family units |
| Detailed participant report |
| Core status EEG/MRI |
| Enrolled IGE-LRE participants by site |
| Enrolled participants by ethnicity race and gender/by type by site |
| Participants lost detail |
| Yearly stats—activities/participants/family units |
2.16. Server and network infrastructure
EPGP uses a redundant, secure, reliable, Internet-connected high-speed network and Virtual Private Network (VPN). Systems are “virtualized” and hosted at the UCSF Secure Datacenter. The center is staffed 24 h a day and newly built to withstand high seismic activity with redundant cooling systems, uninterruptible power supply and diesel generated power backup. In the datacenter, the EPGP servers reside in VMware clusters for high availability and redundancy. There is no single point of failure. This architecture provides a highly flexible and scalable architecture for EPGP computing services.
2.17. Data management
The EPGP data manager application automates most of the data management tasks performed for the EPGP study. These tasks include updating the status of participants’ schedule of events/activities as phenotyping activities are completed and starting EEG and MRI workflows.
2.18. Informatics/IT technical support
The informatics team assists PIs and study coordinators to use the web-based applications, and answers any technical enquiries via telephone and email support. Informatics services include data management, software maintenance, scheduled one-to-one training, user manuals, and on-demand web-based training videos. Data management services include manually editing data when requested and generating one-off data extracts. The data management team also processes EEG and MRI files, which entails removing any personally identifiable information from the source files and converting these source files (e.g. EEG data files) from vendor-specific formats to accessible formats.
2.19. Security and privacy
Each participant is assigned a unique Participant Identifier (alphanumeric string) that is used to identify the participant’s phenotypic records and specimens. All software applications and systems use encryption technologies and conform to all 21CFR Part 11 and HIPAA requirements. The web-based applications use SSL 128 bit encryption and users are forced to input their unique username and password before gaining access to any of the applications. All updates made to the data in the database are tracked and auditable. No protected health information is stored in the database, and participants are tracked using their unique Participant Identifier only.
3. Results
3.1. Phenotypic data collection
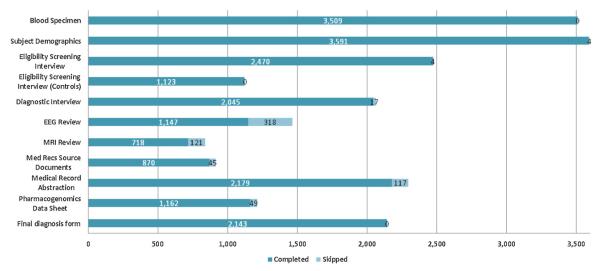
As of January 26th 2012, EPGP has successfully enrolled 3780 study participants. Of the 27,770 activities expected to be carried out for these participants, 21,632 (78%) activities have been completed. Fig. 6 shows the number of completed activities, which includes blood specimens stored at Coriell’s repository, web-based data collection forms, medical record source documents uploaded, EEG reviews and MRI reviews. This represents over 3 million completed data-points in the EPGP data warehouse. 2258 EEGs (a combination of digital EEGs and EEG reports in PDF format) have been uploaded and consume over 60GB of file storage. 1470 MRIs (compressed DICOM files) have been uploaded and consume over 50GB of file storage. EPGP has collected complete AED histories on 1162 study participants. The AED Core has reviewed 764 (66%) of these participants’ AED data thus far. The Data Review Core has reviewed 471 participants’ data, the purpose of which is to identify potential corrections to the phenotypic data and enhance the quality of the data collected.
Fig. 6.
EPGP study activities completed.
3.2. End user management
There are approximately 100 active end-users in EPGP’s database, mainly comprised of study coordinators, PIs/Co-PIs and administrative/IT staff. End-users are provided with at least one hour of one-to-one training to use EPGP’s informatics platform. We provide end-user support via a toll-free telephone number and email, and we receive between 50 and 100 end-user support requests each week. Support requests typically consist of new user account requests, requests for training, and data management requests.
3.3. End-user satisfaction
According to Doll and Torkzadeh [15], there are five components of user satisfaction with information systems: content, accuracy, format, ease of use, and timeliness. Using their validated instrument for measuring end-user satisfaction, we conducted a survey of 85 end-users (21 PIs and 64 study coordinators only) about their level of satisfaction with EPGP’s suite of web-applications. The end-user satisfaction construct was developed with a five point Likert-type scale (1 = almost never; 2 = some of the time; 3 = about half of the time; 4 = most of the time; and 5 = almost always). The response rate was 48% (11 PIs and 30 study coordinators), which is consistent with information systems management research [16]. As shown in Table 2, the results of the survey strongly suggest that the level of end-user satisfaction is very high across all five components and there is little difference in satisfaction between the two groups. The level of end-user satisfaction of PIs was higher than those of study coordinators for content, accuracy and timeliness. Study coordinators’ level of satisfaction was higher than those of PIs for format and ease of use.
Table 2.
End-User satisfaction survey results summary.
| n = 41 | Principal investigator (PI) (n = 11) |
Study coordinator (n = 30) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Question | Mode | Median | Average | SD | ≥4-Most of the time | Mode | Median | Average | SD | ≥4-Most of the time |
| G1. Is the system successful? | 5 | 5 | 4.5 | 0.82 | 82% | 5 | 5 | 4.6 | 0.50 | 100% |
| G2. Are you satisfied with the system? | 4 | 4 | 4.2 | 0.87 | 91% | 5 | 5 | 4.4 | 0.85 | 90% |
| C1. Does the system provide the precise information you need? |
4 | 4 | 4.2 | 0.87 | 91% | 5 | 5 | 4.4 | 0.73 | 87% |
| C2. Does the information content meet your needs? |
4 | 4 | 4.2 | 0.87 | 91% | 5 | 5 | 4.5 | 0.57 | 97% |
| C3. Does the system provide reports that seem to be just about exactly what you need? |
4 | 4 | 4.0 | 0.63 | 82% | 5 | 4 | 4.1 | 0.98 | 70% |
| C4. Does the system provide sufficient information? |
4 | 4 | 4.2 | 0.60 | 91% | 5 | 5 | 4.5 | 0.68 | 90% |
| A1. Is the system accurate? | 5 | 5 | 4.5 | 0.69 | 91% | 5 | 5 | 4.4 | 0.93 | 83% |
| A2. Are you satisfied with the accuracy of the system? |
4 | 4 | 4.1 | 0.83 | 91% | 5 | 5 | 4.4 | 0.85 | 90% |
| F1. Do you think the output is presented in a useful format? |
4 | 4 | 3.9 | 0.83 | 82% | 5 | 5 | 4.3 | 0.92 | 90% |
| F2. Is the information clear? | 5 | 5 | 4.4 | 0.81 | 82% | 5 | 5 | 4.4 | 0.77 | 90% |
| E1. Is the system user friendly? | 5 | 4 | 3.9 | 1.30 | 64% | 5 | 5 | 4.4 | 0.85 | 83% |
| E2. Is the system easy to use? | 4 | 4 | 4.1 | 0.94 | 82% | 5 | 5 | 4.4 | 0.81 | 87% |
| T1. Do you get the information you need in time? |
4 | 4 | 4.2 | 0.60 | 91% | 5 | 5 | 4.5 | 0.68 | 90% |
| T2. Does the system provide up-to-date information? |
5 | 5 | 4.4 | 0.81 | 82% | 5 | 5 | 4.3 | 0.94 | 73% |
3.4. File transfer
EEGs and MRIs can be uploaded and downloaded extremely quickly and securely using FTP software. In a simple test, a 27MB EEG file was down loaded from EPGP’s FTP server in less than 50 s over a 15 Mb/s internet download connection. Most clinical sites have even faster connections to the internet, so can achieve even greater download speeds, therefore making this a very effective and efficient alternative to sending physical CD/DVDs using costly traditional methods.
4. Discussion
Though academic research projects are less likely to have IT support solutions when compared to industry-sponsored research projects [17], we designed and implemented a suite of novel web-based informatics tools for data collection and storage, specimen tracking, and data review. The successful implementation of the EPGP informatics platform has streamlined and standardized data collection methods, enabling us to manage the activities for the entire study in real-time and maximizing the quality of the phenotypic data collected.
EPGP’s informatics platform enabled us to conduct phenotyping activities and to collect large amounts of phenotypic data using web-based tools, resulting in substantial improvements and efficiencies over paper-based approaches [18]. The effective use of electronic data-capture tools helped ensure high-quality data were available for early review and rapid decision-making [19].
End-users that responded to the satisfaction survey are, for the most part, very satisfied. The results of the end-user satisfaction survey suggested that study coordinators found the web-based applications easy to use and that information was displayed in a clear and useful format, and that PIs were provided with precise and accurate information in a timely manner.
Through there are other off-the-shelf applications available to help PIs set-up and manage their clinical studies, such as REDCap [20] and OpenClinica [21], we found that none of them adequately met the needs of EPGP. EPGP is somewhat unique in that the study was conducting “deep phenotyping” on its participants, which required the use of very lengthy data collection forms that had extremely complex branching (skip logic) requirements and question types. As the scientific needs of the EPGP study evolved during the early stages of the study, the informatics platform had to have the ability to change quickly too, and we would not have been able to achieve this agility or control with an off-the-shelf informatics platform.
A web-based system to manage and coordinate multisite studies is essential [22,23], and EPGP has benefited from its custom developed informatics platform. Some of our future plans include redesigning the database architecture and applications so that they can accommodate multiple studies concurrently and can be easily adapted to suit the specific requirements of these studies, without entailing any significant software changes. The experience gained from implementing and supporting the EPGP informatics platform will help with the design of new informatics tools to support future large scale collaborative research studies, and will, through this experience, help advance the domain of clinical research informatics [24].
Summary points.
What was known before
There are no pre-existing informatics platforms that could handle the combination of data points that we have in EPGP, especially EEG and MRI, and the approach for phenotype verification that requires a Data Review Core.
What the study has added
A web-based system to manage and coordinate multisite studies is essential.
EPGP has benefited from its custom developed informatics platform.
The quality of the data collected has been improved through real-time access to the data, built-in data validation in the web-based data collection forms and by web-based data review applications.
The EPGP informatics platform, and the experience gained developing it, will help with the design of new informatics tools to support future clinical research studies.
Acknowledgments
Grant support This work is supported by National Institute of Neurological Diseases and Stroke (NINDS) grant U01 NS053998, as well as planning grants from the Finding a Cure for Epilepsy and Seizures Foundation and the Richard Thalheimer Philanthropic Fund. We would like to acknowledge the recruitment contributions of the EPGP Community Referral Network (CRN). The CRN consists of healthcare professionals not paid by the EPGP grant who refer eligible families to EPGP. A list of individual contributors can be found at www.epgp.org. In addition, we would like to acknowledge the efforts of the clinical coordinators, the site principal investigators, neurologists, and support staff at our EPGP clinical centers who have contributed significant effort into recruitment, data acquisition and storage, and extensive phenotyping. Finally, we extend our sincere appreciation to the participants with epilepsy and their families who have contributed to this research effort.
Appendix A. Contributors to informatics platform design
Informatics core members
Gerry Nesbitt, MBA, PMP, Director of Informatics
Kevin McKenna, Database Manager
Vickie Mays, Data Coordinator
Alan Carpenter, Programmer Analyst
Kevin Miller, Programmer Analyst
Michael Williams, Informatics CIO
EPGP investigators
Bassel Abou-Khalil, MD; Data Review Core, Local PI; Vanderbilt University Medical Center
Brian Alldredge, PharmD; AED Core; University of California, San Francisco
Jocelyn Bautista, MD; Local PI; Cleveland Clinic
Sam Berkovic, MD; Local PI; The University of Melbourne
Alex Boro, MD; EEG Core; Albert Einstein College of Medicine Gregory Cascino, MD; MRI Core, Local PI; Mayo Clinic College of Medicine Rochester, Minnesota
Damian Consalvo, MD, PhD; Local PI; Hospital General de Agudos José Maria Ramos Mejía
Patricia Crumrine, MD; Local PI; Children’s Hospital of Pittsburgh of UPMC
Orrin Devinsky, MD; Phenotyping Core, Local PI; New York University School of Medicine
Dennis Dlugos, MD, MCSE; EEG Core, Phenotyping Core, Local PI; The Children’s Hospital of Philadelphia
Michael Epstein, PhD; Data Analysis Core; Emory University School of Medicine
Miguel Fiol, MD; Referral Center PI; University of Minnesota Medical Center
Nathan Fountain, MD; Data Review Core, Local PI; University of Virginia Health System
Jacqueline French, MD; AED Core; New York University School of Medicine
Daniel Friedman, MD; Local Co-PI; New York University School of Medicine
Eric Geller, MD; Local Co-PI; St. Barnabas Health Care System
Tracy Glauser, MD; AED Core, Local PI; Cincinnati Children’s Hospital Medical Center
Simon Glynn, MD; Local PI; University of Michigan
Sheryl Haut, MD, MS; Local PI; Albert Einstein College of Medicine
Jean Hayward, MD; Referral Center PI; Kaiser Permanente: Oakland Medical Center
Sandra Helmers, MD; Local PI; Emory University School of Medicine
Andres Kanner, MD; AED Core; Rush University Medical Center
Heidi Kirsch, MD, MS; Local PI; University of California, San Francisco
Robert Knowlton, MD; MRI Core, Local PI; University of Alabama at Birmingham School of Medicine
Eric Kossoff, MD; Local Co-PI; The Johns Hopkins University School of Medicine
Rachel Kuperman, MD; Local Referral Center PI; Children’s
Hospital & Research Center Oakland
Ruben Kuzniecky, MD; Study PI; New York University School of Medicine
Daniel Lowenstein, MD; Study PI; University of California, San Francisco
Shannon McGuire, MD; Local PI; Louisiana State University Health Sciences Center
Paul Motika, MD; Local Co-PI; Rush University Medical Center
Edward Novotny, MD; Local PI; Seattle Children’s Hospital
Ruth Ottman, PhD; Phenotyping Core; Columbia University
Juliann Paolicchi, MD; Local PI; Vanderbilt University Medical Center
Jack Parent, MD; Local Co-PI; University of Michigan
Kristen Park, MD; Local PI; The Children’s Hospital Denver
Annapurna Poduri, MD; Data Review Core, Local PI; Children’s Hospital Boston
Neil Risch PhD; Data Analysis Core; University of California, San Francisco
Lynette Sadleir, MBChB, MD; Local PI; Wellington
School of Medicine and Health Sciences, University of Otago
Ingrid Scheffer, MBBS, PhD; Data Review Core, Local PI; The University of Melbourne
Renee Shellhaas, MD; EEG Core; University of Michigan
Elliot Sherr, MD, PhD; Phenotyping Core; University of California, San Francisco
Jerry Shih, MD; Data Review Core, Local PI; Mayo Clinic College of Medicine Jacksonville, Florida
Shlomo Shinnar, MD, PhD; Phenotyping Core; Albert Einstein College of Medicine
Rani Singh, MD; Local Co-PI; University of Michigan
Joseph Sirven, MD; Local PI; Mayo Clinic College of Medicine Scottsdale, Arizona
Michael Smith, MD; Local PI; Rush University Medical Center
Joe Sullivan, MD; EEG Core; University of California, San Francisco
Liu Lin Thio, MD, PhD; Local PI; Washington University in St. Louis
Anu Venkatasubramanian, MD; Local Co-PI; The Children’s Hospital of Philadelphia
Eileen Vining, MD; Local PI; The Johns Hopkins University School of Medicine
Gretchen Von Allmen, MD; Local PI; University of Texas Health Science Center at Houston
Judith Weisenberg, MD; Local PI; Washington University in St. Louis
Peter Widdess-Walsh, MD; Local PI and Data Review Core; St. Barnabas Health Care System
Melodie Winawer, MD, MS; Phenotyping Core and Data Review Core; Columbia University
Administrative core members
Catharine Freyer, Project Director
Kristen Schardein, RN, MS, Recruitment Director
Robyn Fahlstrom, MPH, Statistician
Sabrina Cristofaro, RN, BSN, Phenotyping Director
Kathleen McGovern, Recruitment Assistant
Nora Stillman, Recruitment Assistant
Study coordinators
PrashantAgarwal; Study Coordinator; The Children’s Hospital of Philadelphia
Jennifer Ayala; Study Coordinator; Albert Einstein College of Medicine
Cate Bakey; Study Coordinator; The Children’s Hospital of Philadelphia
Thomas Borkowski, PhD; Study Coordinator; Albert Einstein College of Medicine
Riann Boyd, CCRP; Study Coordinator; Mayo Clinic College of Medicine Jacksonville, Florida
Alicia Camuto, CCRP; Study Coordinator; The Children’s Hospital Denver
Cendy Carrasco; Study Coordinator; University of California, San Francisco
Jennifer Cassarly; Study Coordinator; Cincinnati Children’s Hospital Medical Center
Yong Collins, RN; Study Coordinator; The Children’s Hospital of Philadelphia
Kevin Collon; Study Coordinator; University of Michigan
Sean Collon; Study Coordinator; University of Michigan
Heather Eckman; Study Coordinator; New York University School of Medicine
Susan Fogarty, RN; Study Coordinator; University of Texas
Health Science Center at Houston
Dolores González Morón, MD; Study Coordinator; Hospital General de Agudos José Maria Ramos Mejía
La June Grayson; Study Coordinator; Washington University in St. Louis
Samantha Hagopian, RN; Study Coordinator; The Children’s Hospital of Philadelphia
Emily Hayden; Study Coordinator; University of Michigan
Kristin Heggeli; Study Coordinator; Mayo Clinic College of Medicine Jacksonville, Florida
Rachel Hennessy; Study Coordinator; New York University School of Medicine
Jody Hessling, RN; Study Coordinator; Cincinnati Children’s Hospital Medical Center
Emily Hirschfield; Study Coordinator; Cincinnati Children’s Hospital Medical Center
Jennifer Howell, BA, BS; Study Coordinator; University of Alabama at Birmingham School of Medicine
Sherry Klingerman; Study Coordinator; Mayo Clinic College of Medicine Rochester, Minnesota
Maritza Lopez, RN; Study Coordinator; University of California, San Francisco
Heather Marinelli; Study Coordinator; The Johns Hopkins University School of Medicine
Brandy Maschhaupt; Study Coordinator; Mayo Clinic College of Medicine Scottsdale, Arizona
Jennie Minnick; Study Coordinator; The Children’s Hospital of Philadelphia
Jade Misajon, RN; Study Coordinator; St. Barnabas Health Care System
Jennifer Monahan, RN; Study Coordinator; Children’s Hospital of Pittsburgh of UPMC
Karen Oliver; Study Coordinator; The University of Melbourne
Isha Parulkar; Study Coordinator; Children’s Hospital Boston
Laura Przepiorka, RN; Study Coordinator; Rush University Medical Center
Paula Pyzik, BA, CRT; Study Coordinator; The Johns Hopkins University School of Medicine
Brigid Regan; Study Coordinator; The University of Melbourne
Catherine Shain; Study Coordinator; Children’s Hospital Boston
Lexie Slingerland; Study Coordinator; The University of Melbourne
Caitlin Stanton, MPH; Study Coordinator; Seattle Children’s Hospital
Kelly Taylor, MS, CGC; Study Coordinator; Vanderbilt University Medical Center
Stacy Thompson, RN, BSN, CCRC; Study Coordinator; University of Virginia Health System
Jennifer Turczyk; Study Coordinator; Cleveland Clinic
Alexander Vara; Study Coordinator; University of Texas Health Science Center at Houston
Cindy Wesolowski, RN, NP; Study Coordinator; Cincinnati Children’s Hospital Medical Center
Andrew Yourich; Study Coordinator; University of Alabama at Birmingham School of Medicine
Footnotes
Author contributions All authors qualify for authorship by substantial contributions to the research and production of the manuscript. Gerry Nesbitt was chief architect of the design of the informatics applications, compiled the initial draft of this report. Kevin McKenna provided significant input into the data management aspects of the informatics platform and Vickie Mays provided significant input with regards to the processing of EEGs, MRIs and medical records, and both revised the draft critically prior to submission. Alan Carpenter and Kevin Miller provided input into building the Informatics platform. Michael Williams formed the Informatics and IT teams, and contributed to the overall technical vision, architecture and solution. All authors gave final approval for the submitted manuscript.
Competing interests All authors declare that they have no conflicts of interest, financial or otherwise to disclose.
REFERENCES
- [1].Epilepsy Phenome/Genome Project [last accessed 14.07.11]; Available from: http://www.epgp.org.
- [2].Pavlovića I, Kern T, Miklavčiča D. Comparison of paper-based and electronic data collection process in clinical trials: costs simulation study. Contemp. Clin. Trials. 2009 Jul 4;30:300–316. doi: 10.1016/j.cct.2009.03.008. [DOI] [PubMed] [Google Scholar]
- [3].National Institutes of Health [last accessed 14.07.11];The Health Insurance Portability and Accountability Act of 1996 (HIPAA) Privacy and Security Rules. Available from: http://privacyruleandresearch.nih.gov.
- [4].US Food & Drug Administration [last accessed 14.07.11];CFR—Code of Federal Regulations Title 21. Available from: http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/cfrsearch.cfm?cfrpart=11.
- [5].Epilepsy Phenome/Genome Project [last accessed: 19.01.12];Eligibility Criteria. from: http://www.epgp.org/Pages/Eligibility.aspx.
- [6].Coriell Institute for Medical Research. Camden, NJ: [last accessed: 14.07.11]. Available from: http://www.coriell.org. [Google Scholar]
- [7].Franklin J, Guidry A, Brinkley J. A partnership approach for Electronic Data Capture in small-scale clinical trials. J. Biomed. Inform. 2011 May 30; doi: 10.1016/j.jbi.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. [last accessed 14.07.11];Microsoft Sharepoint. 2010 Available from:http://sharepoint.microsoft.com.
- [9]. [last accessed 14.07.11];Microsoft Silverlight. Available from:http://www.microsoft.com/silverlight.
- [10]. [last accessed 14.07.11];International League Against Epilepsy (ILAE) Available from:http://www.ilae-epilepsy.org.
- [11]. [last accessed 14.07.11];Microsoft SQL Server. 2008 Available from:http://www.microsoft.com/sqlserver.
- [12].Persyst Development Corporation. Prescott, AZ: [last accessed 14.07.11]. Available from: http://www.eeg-persyst.com. [Google Scholar]
- [13].National Electrical Manufacturers Association (NEMA) [last accessed 14.07.11];Digital Imaging and Communications in Medicine (DICOM) Available from: http://www.nema.org/stds/dicom.cfm.
- [14].Nesbitt G, Carpenter A, Dlugos D, Sullivan J, Shellhaas R, Boro A, et al. EPGP: Informatics Tools and Workflow for Processing Electroencephalogram (EEG) Data. American Epilepsy Society. 2010 Abst. 2.047. [Google Scholar]
- [15].Doll WJ, Torkzadeh G. The measurement of end-user computing satisfaction. MIS Quart. 1988;12(2):259–275. [Google Scholar]
- [16].Falconer DJ, Hodgett RA. Why executives don’t respond to your survey. Proceedings of the Australasian Conference on Information Systems; Wellington, NZ. 1999. [Google Scholar]
- [17].EI Emam K, Jonker E, Sampson M, et al. The use of electronic data capture tools in clinical trials: web-survey of 259 Canadian trials. J. Med. Internet Res. 2009;11:e8. doi: 10.2196/jmir.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Marks RG, Conlon M, Ruberg SJ. Paradigm shifts in clinical trials enabled by information technology. Stat. Med. 2001 Sep 17–18;20:2683–2696. doi: 10.1002/sim.736. [DOI] [PubMed] [Google Scholar]
- [19].Lu Z, Su J. Clinical data management: Current status, challenges, and future directions from industry perspectives. Open Access J. Clin. Trials. 2010 Jun 2;:93–105. 2010. [Google Scholar]
- [20].Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009 Apr 2;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. [last accessed 14.07.11];Open Clinica. Available from: http://www.openclinica.org.
- [22].Winget M, Kincaid H, Lin P, Li L, Kelly S, Thornquist M. A web-based system for managing and coordinating multiple multisite studies. Clin. Trials. 2005 Feb 1;2:42–49. doi: 10.1191/1740774505cn62oa. [DOI] [PubMed] [Google Scholar]
- [23].Lallas CD, Preminger GM, Pearle MS, Leveillee RJ, Lingeman JE, Schwope JP, Pietrow PK, Auge BK. Internet based multi-institutional clinical research: a convenient and secure option. J. Urol. 2004 May 5;171:1880–1885. doi: 10.1097/01.ju.0000120221.39184.3c. [DOI] [PubMed] [Google Scholar]
- [24].Embi P, Payne P. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J. Am. Med. Inform. Assoc. 2009 May-Jun;16:316–327. doi: 10.1197/jamia.M3005. [DOI] [PMC free article] [PubMed] [Google Scholar]