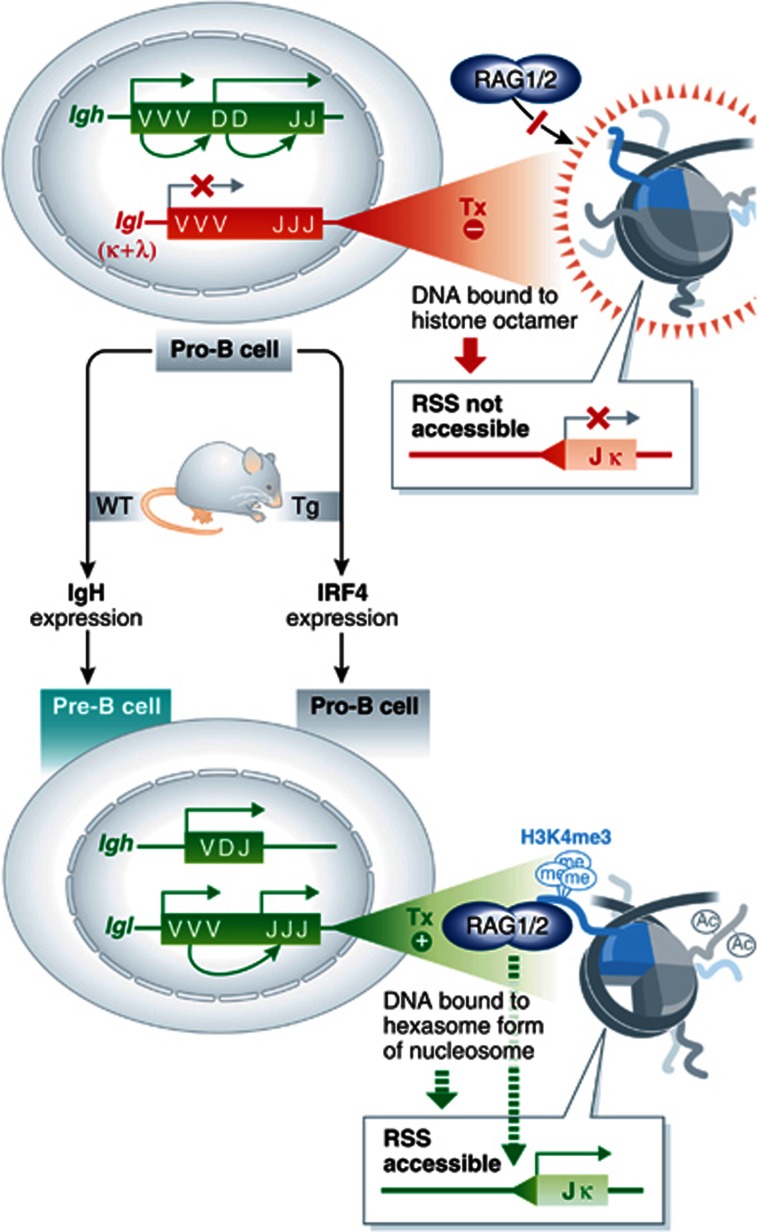
Figure 1.
(Left) Developmental control of Ig gene rearrangements. Pro-B cells normally undergo Igh recombination (active, green), but repress assembly of Igk and Igl light chain genes (inactive, red). Upon functional assembly and expression of Igh, wild-type (WT, left arrow) pro-B cells differentiate into pre-B cells and activate Igk/l recombination. Bevington and Boyes show that the developmental block in Igk/l activation can be overcome by transgene-driven expression of IRF4 in pro-B cells (Tg, right arrow). (Right) Authors’ model of recombinase accessibility. RSSs (triangle) flanking Igk/l gene segments are transcriptionally inert (Tx –) in wild-type pro-B cells (top) and associate with conventional nucleosome octamers, which block access to the RAG complex. In pre-B cells or pro-B cells expressing IRF4, transcriptional activation decorates histone tails with acetylation (Ac) and H3K4me3, the latter of which serves as a platform for stable docking of RAG complexes. Transcription also leads to the expulsion of histone dimers, resulting in a hexasome form of nucleosomes, which may unmask RSSs for cleavage by RAG.