Fig. 4.
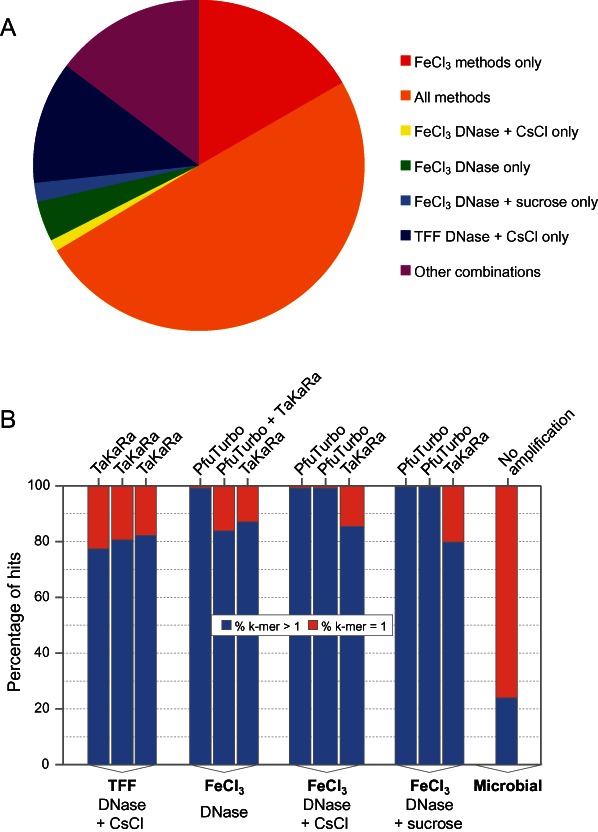
K-mer-based analysis of shared reads between methods and replicates. (A) Percentage of total reads that are shared between methods based on a k-mer-based analysis and (B) percentage of ‘rare’ (k-mer = 1) versus abundant sequences (k-mer > 1) in each of the four viral metagenomic methods and a non-replicated microbial metagenome. The enzymes used for linker amplification (TaKaRa or PfuTurbo) are listed above each sample. The microbial sample includes more ‘rare’ sequences because the diversity in the sample is undersampled based upon rarefaction analysis (data not shown).
