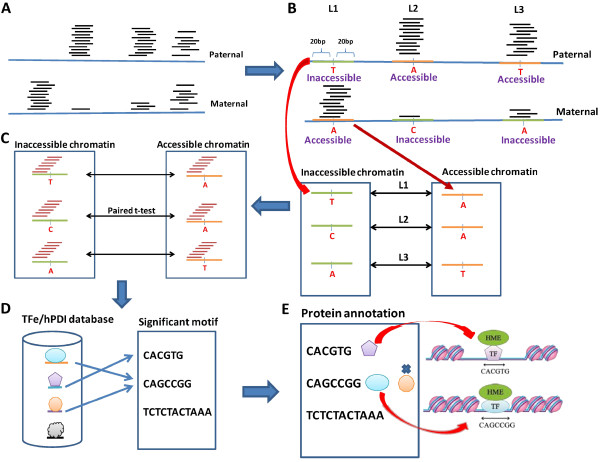
Figure 1.
Overview of the algorithm. (A) To identify accessible and inaccessible chromatin regions, we mapped FAIRE-seq reads to a diploid genome (GM12878) using Bowtie without any mismatch. (B) We identified differential chromatin-state regions (DCSRs) and then separated DCSRs into accessible and inaccessible chromatin groups. (C) To discover chromatin motifs, we used all combinations of DNA sequences from 6mer to 10mer (motif candidates) to scan all of the DCSRs, each of which has 41 bases in total. For each motif candidate, we calculated occurrence frequency, and then performed paired t-tests between frequency vectors of the inaccessible chromatin group and accessible chromatin group. We selected chromatin motifs with P values < 0.01 by the paired t-tests. (D) To annotate the motifs, we used the motif sequences and then performed BLAST alignment against the entire transcription factor binding site sequences in both the TFe and hPDI databases. For each motif, we annotated the motif using the best E-value of alignment and homology > 0.8. (E) To provide more accurate annotation, we filtered out the transcript factors that are not expressed (FPKM < 1) by the RNA-seq data (SRP007417).