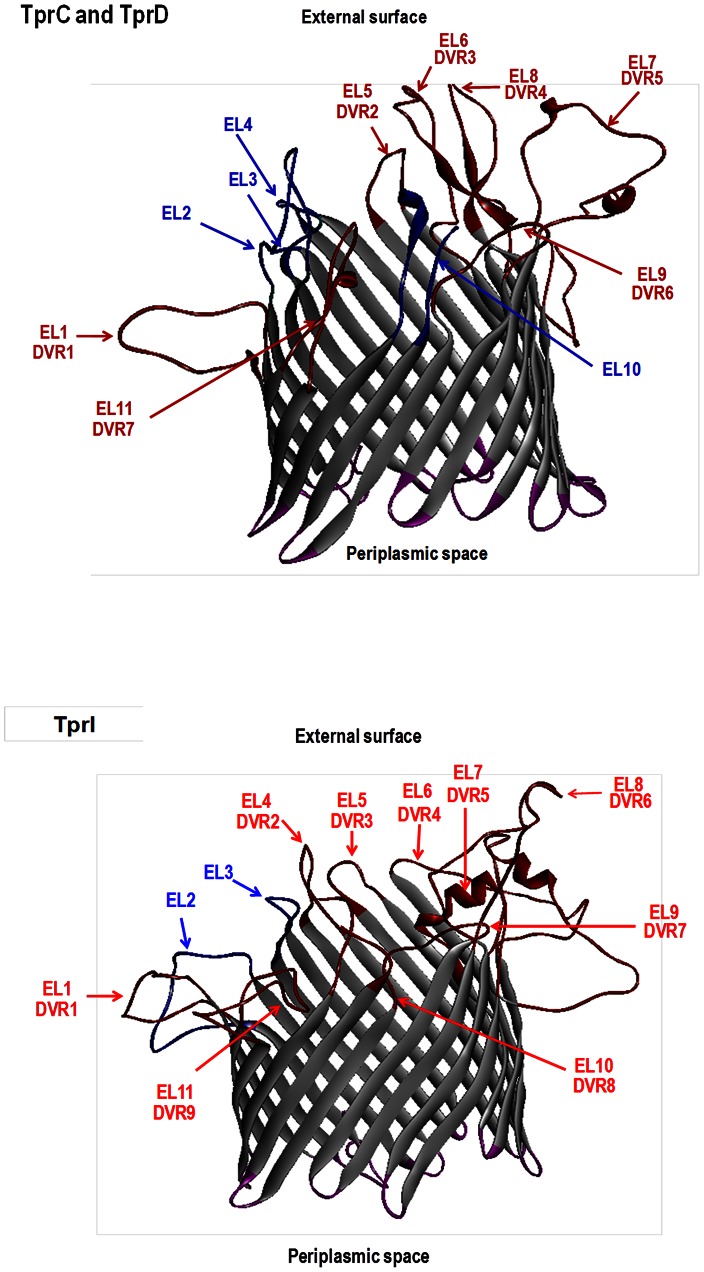
Figure 3. Structural models of TprC/D and TprI.
Non-templated 3D models generated for the mature Nichols TprC/D and Nichols TprI peptides using the TMBpro algorithm [61] suggest a typical β-barrel structure. DVR,discrete variable regions. EL, external loops. Variable regions, DVR1–DVR7 for TprC/D and DVR1–DVR9 for TprI, as defined by protein sequence alignments (Figure S1 and S2) are indicated by red color (loops, font and arrows). Note that each DVR co-localizes with a predicted EL. Orientation of the structure was determined as specified by Randall et al [61]. Proposed conserved and variable surface exposed loops are highlighted in blue and red, respectively, and proposed periplasmic exposed regions of the proteins are in purple.