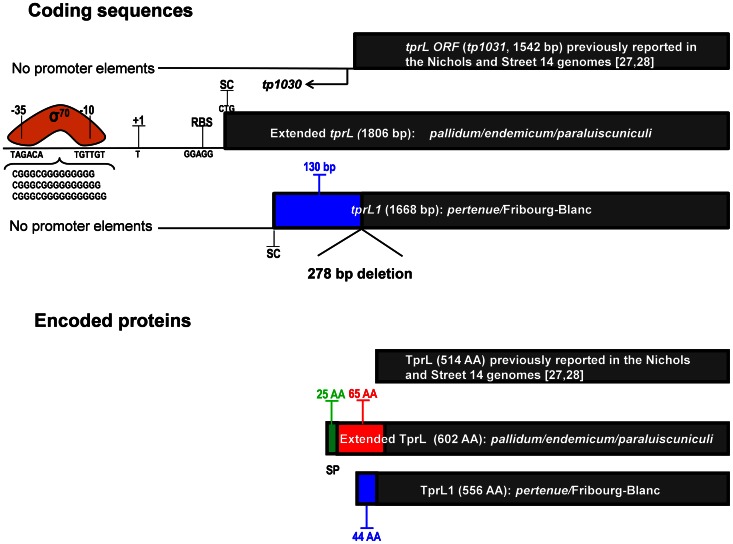
Figure 6. Encoded variants at the tprL (tp1031) locus.
Coding sequences: Three different coding sequences have been identified for treponemal species and subspecies: the proposed tprL ORFs in the Nichols and Street 14 genome sequences; an extended tprL for pallidum, endemicum, and paraluiscuniculi strains; and a fused tprL (called tprL1) for pertenue and the Fribourg-Blanc strains. The Nichols ORF was predicted to be 1542 bp, although lacks identifiable promoter elements upstream. In this study, an extended tprL of 1806 bp has been identified in the Nichols and other pallidum strains, as well as in endemicum and paraluiscuniculi strains. The initially shorter Nichols tprL was the result of sequencing errors in the reported Nichols genome sequence [27]. Typical promoter elements are shown for the extended tprL ORF (SC, start codon. RBS, ribosomal binding site. +1, transcriptional start site (TSS). −10 and −35, σ70 signatures). A deletion of 278 bp (274 bp of the 5′ end of tp1030, whose coding sequence is located on the minus strand, and 4 bp of the 5′ end of the genome-derived tprL) creates an alternative start site in tp1030 for pertenue and Fribourg-Blanc tprL1, resulting in a shorter ORF of 1668 base pairs. This ORF, however, lacks recognizable promoter elements. Encoded proteins: Differences in coding sequences result in two different proteins: 1) a shorter pertenue/Fribourg-Blanc variant with a 44 amino acid unique amino terminus and 2) a longer TprL in the remaining species/subspecies with a predicted signal peptide 25 amino acids long (green) in the longer product, but not identifiable in the pertenue/Fribourg-Blanc gene product. Blue color, region unique to pertenue and Fribourg-Blanc strains (132 nucleotides or 44 amino acids). Red color, region unique to the pallidum, endemicum and paraluiscuniculi species/subspecies (65 amino acids).