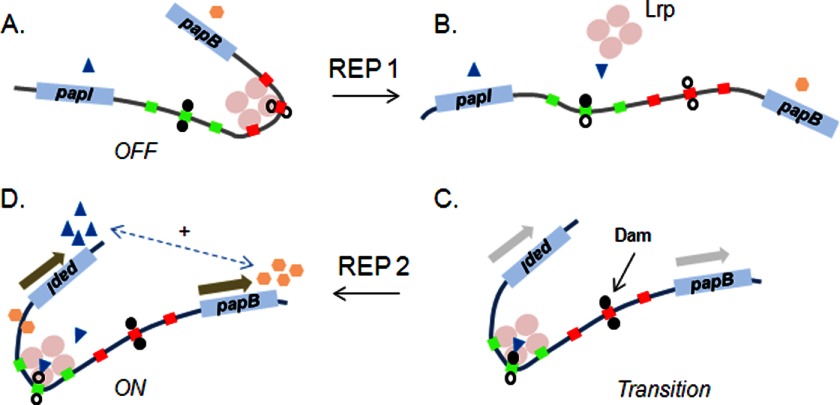
FIGURE 3.
Model for the pap off-to-on state transition, an example of a bistable switch controlled by DNA methylation patterns. A, in the phase off state, an octamer of Lrp (a tetramer of dimers; only one tetramer is depicted) binds cooperatively to promoter-proximal sites 1–3 (red boxes). Lrp binding to sites 1–3 inhibits further binding of Lrp to sites 4–6 (green boxes) by mutual exclusion. B, immediately following passage of the replication fork (REP 1), the two daughter chromosomes become hemimethylated. Only the daughter chromosome methylated on the top strand is shown (filled circle above Lrp-binding site 5). C, two stochastic events occur in which PapI facilitates Lrp binding to sites 4–6, and Dam (DNA adenine methylase) methylates both strands of the proximal GATC site. Binding of Lrp at sites 4–6 reduces the affinity of Lrp for sites 1–3 by mutual exclusion and facilitates activation of pap transcription via cAMP-catabolite gene activator protein/RNA polymerase binding (not shown). Methylation of GATCprox reduces the affinity of PapI/Lrp for sites 1–3 and is required for transition to the on phase (98). D, one additional round of DNA replication (REP 2) completes transition to the phase on state, in which GATCdist is fully unmethylated. The on phase is self-perpetuating due to a bidirectional feedback loop between PapB and PapI (dashed arrow). The PapB level rises due to activation of transcription of the first gene of the pap operon, papB. PapB binds near the papI promoter, increases the PapI level via activation of papI transcription, and helps maintain the on state via binding of PapI/Lrp to sites 4–6.