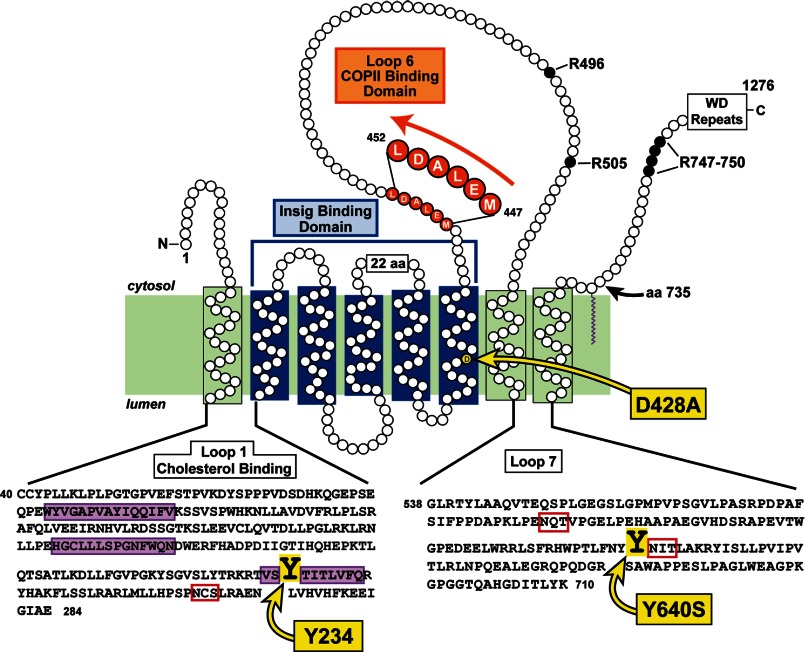
FIGURE 1.
Topology model of the membrane domain of hamster Scap, showing its three functional domains and the sites of three point mutations (Y234A, D428A, and Y640S) that confer a constitutive cholesterol-bound conformation, even in the absence of sterols. Amino acids (aa) 40–284 correspond to the sequence of luminal Loop 1, the cholesterol-binding domain of Scap. The three hydrophobic patches in the Loop 1 sequence are shaded in purple, and the N-linked glycosylation site is denoted by the red box. The Insig-binding domain is localized to transmembrane helices 2–6, shown by the blue bracket. The COPII-binding site is localized to the MELADL sequence in Loop 6, shaded in orange. Amino acids 538–710 correspond to the sequence of luminal Loop 7; its two N-linked glycosylation sites are denoted by the red boxes. In membranes from sterol-deprived cells, trypsin cleaves Scap on its NH2-terminal side at Arg-496; in sterol-replete membranes, trypsin cleaves at Arg-503/Arg-505. The trypsin-cleavage site on the COOH-terminal side of Scap in both the absence and the presence of sterols occurs within a cluster of arginines (Arg-747–Arg-750).