FIGURE 2.
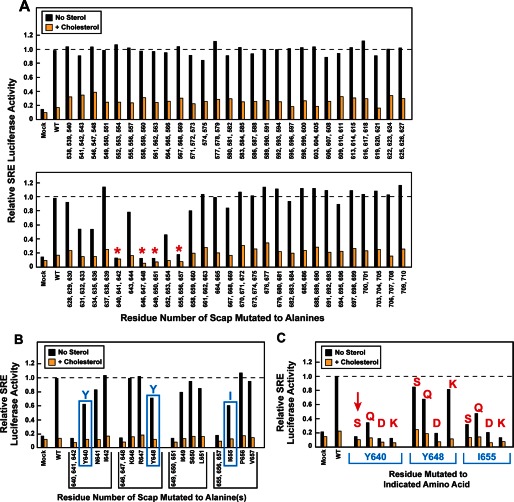
Alanine-scanning mutagenesis of Loop 7 region of hamster Scap. On day 0, SRD-13A cells were set up for experiments in 1 ml of medium A containing 5% FCS at a density of 2.5 × 104 cells/well in 24-well plates. On day 1, cells were co-transfected in 1 ml of medium A supplemented with 5% FCS containing 38 ng of pTK-Insig1-Myc, 125 ng of pSRE-firefly luciferase, 125 ng of pTK-Renilla luciferase, and 50 ng of WT or the indicated mutant pTK-Scap in which 2–3 contiguous residues were mutated to alanine. FuGENE 6 was used as the transfection agent. For each transfection, the total amount of DNA was adjusted to 338 ng/dish by the addition of pcDNA mock vector. On day 2 (after incubation with plasmids for 24 h), the cells were washed once with PBS and then treated with hydroxypropyl-β-cyclodextrin-containing medium B for 1 h. Cells were then washed twice with PBS and re-fed with 1 ml of medium C in the absence or presence of 15 μm cholesterol complexed with methyl-β-cyclodextrin. After incubation for 16 h, the cells were washed with PBS, after which luciferase activity was read on a Synergy 4 plate reader (BioTek) according to the Promega protocol. The amount of SRE-firefly luciferase activity in each dish was normalized to the amount of Renilla luciferase activity in the same dish. Relative SRE-luciferase activity of 1.0 represents the normalized luciferase value in dishes transfected with WT pTK-Scap in the absence of cholesterol. All values are the average of duplicate assays. A, red asterisks denote triple contiguous alanine mutations that produced a loss of SRE-luciferase activity in both the absence and the presence of cholesterol. The data in these graphs were obtained in four different experiments, each with its own WT control. B, deconvolution of triple mutants that showed loss of SRE-luciferase activity in A. Blue boxes denote single alanine mutations that show partial loss of SRE-luciferase activity. C, substituting serine, glutamine, aspartic acid, and lysine in place of Tyr-640, Tyr-648, and Ile-655. The red arrow denotes the Y640S mutation that was selected for further characterization. In B and C, single-letter codes are used to denote amino acids.