Background: Complex I is the largest proton pump in mitochondria, yet its mechanism is unknown.
Results: For the first time, inhibitor-sensitive semiquinone and cluster N6 signals were detected in affinity-purified E. coli complex I.
Conclusion: The semiquinone species is involved in the redox-driven proton-pumping mechanism.
Significance: Our highly pure complex I will help advance the mechanistic study of the protein.
Keywords: Electron Paramagnetic Resonance (EPR), Flavin, Iron-Sulfur Protein, Proton Pumps, Quinones, Complex I, NADH Dehydrogenase, Semiquinone
Abstract
NADH:ubiquinone oxidoreductase (complex I) pumps protons across the membrane using downhill redox energy. The Escherichia coli complex I consists of 13 different subunits named NuoA-N coded by the nuo operon. Due to the low abundance of the protein and some difficulty with the genetic manipulation of its large ∼15-kb operon, purification of E. coli complex I has been technically challenging. Here, we generated a new strain in which a polyhistidine sequence was inserted upstream of nuoE in the operon. This allowed us to prepare large amounts of highly pure and active complex I by efficient affinity purification. The purified complex I contained 0.94 ± 0.1 mol of FMN, 29.0 ± 0.37 mol of iron, and 1.99 ± 0.07 mol of ubiquinone/1 mol of complex I. The extinction coefficient of isolated complex I was 495 mm−1 cm−1 at 274 nm and 50.3 mm−1 cm−1 at 410 nm. NADH:ferricyanide activity was 219 ± 9.7 μmol/min/mg by using HEPES-Bis-Tris propane, pH 7.5. Detailed EPR analyses revealed two additional iron-sulfur cluster signals, N6a and N6b, in addition to previously assigned signals. Furthermore, we found small but significant semiquinone signal(s), which have been reported only for bovine complex I. The line width was ∼12 G, indicating its neutral semiquinone form. More than 90% of the semiquinone signal originated from the single entity with P½ (half-saturation power level) = 1.85 milliwatts. The semiquinone signal(s) decreased by 60% when with asimicin, a potent complex I inhibitor. The functional role of semiquinone and the EPR assignment of clusters N6a/N6b are discussed.
Introduction
NADH:ubiquinone oxidoreductase (complex I: EC 1.6.5.3) is the entry enzyme of the respiratory chain in mitochondria. It couples scalar transfer of electrons from NADH to ubiquinone (UQ)2 with vectorial proton translocation across the inner membrane (1–3). Complex I provides ∼40% of the proton-motive force required for ATP synthesis (4). On the other hand, complex I is a major site of reactive oxygen species generation and is extremely vulnerable to oxidative stress. Accordingly, complex I dysfunction has been implicated in a variety of mitochondrial diseases including heart failure, type 2 diabetes, and several neurodegenerative diseases such as Parkinson disease (5). Mammalian complex I is recognized as one of the most elaborate membrane-bound iron-sulfur proteins with a total mass close to 1,000 kDa, and it is composed of 45 different protein subunits (6), seven of which are encoded by mitochondrial DNA (mtDNA) and the rest of which are encoded by nuclear DNA (7, 8). On the other hand, the bacterial complex I catalyzes the same reaction and harbors the same set of cofactors and consists of only 13–17 subunits (2, 9, 10), but at least 13–14 of them have homologs in the mitochondrial enzyme (9). Taking advantage of its simplicity and ease of genetic manipulation, we have used E. coli complex I as a model system to study the structure and function of complex I.
The recent x-ray crystal structures revealed that complex I has a characteristic L-shaped structure with two distinct domains: a hydrophilic peripheral arm and a transmembrane hydrophobic arm (11, 12). The high resolution, three-dimensional structure of Thermus thermophilus HB-8 complex I defined locations of a noncovalently bound flavin mononucleotide (FMN) and nine iron-sulfur clusters, N1a, N1b, N2, N3, N4, N5, N6a, N6b, and N7, in the peripheral arm (13–15). The quinone reduction site is thought to be at the interface between the peripheral and membrane arms. The transmembrane arm is involved in proton pumping (16–20) and quinone (Q)/inhibitor binding (21–26). The mechanism of how electron transfer is linked to vectorial proton translocation, however, remains largely unknown.
Electron paramagnetic resonance (EPR) spectroscopy has been one of the most successful and informative methods to study the properties of iron-sulfur clusters of complex I (27–30), but not all of the clusters are detectable by EPR spectroscopy. For example, if iron-sulfur clusters are not paramagnetic due to their very low midpoint potential, their EPR signals are undetectable. Furthermore, when iron-sulfur clusters have very fast spin relaxation, their EPR signals become too broad to be detected even at extremely low temperatures like 4 K. In addition, considerable overlap of signals exists in the observed EPR spectra for complex I for tetranuclear ([4Fe-4S]) clusters (13, 31). This makes the actual assignment of observed g values to the individual clusters an extremely challenging and debatable issue (14, 32). In E. coli complex I, EPR signals from at least six iron-sulfur clusters (N1a, N1b, N2, N3, N4, and N7) were identified (28–30, 33, 34). Recently, it was suggested that EPR signals from cluster N6a were detectable based on mutational analysis (35).
Here, we developed our new purification system for E. coli complex I by generating a strain in which a polyhistidine sequence was inserted upstream of nuoE in the nuo operon (from NADH:ubiquinone oxidoreductase). This allowed us to prepare large amounts of highly pure and active enzyme by rapid and efficient affinity purification despite its low abundance in the membranes. Our detailed EPR analyses including power saturation profiles and simulation revealed two additional iron-sulfur cluster signals, presumably clusters N6a and N6b, in addition to previously assigned signals. Furthermore, we successfully detected small but significant and inhibitor-sensitive semiquinone (SQ) signal(s), which until now have been reported only for bovine complex I (36, 37). The functional role of SQ in proton-pumping mechanism and the EPR assignment of clusters N6a and N6b are discussed.
EXPERIMENTAL PROCEDURES
Materials
The pCRScript cloning kit, QuikChange® II XL site-directed mutagenesis kit, and Herculase®-enhanced DNA polymerase were obtained from Stratagene (Cedar Creek, TX). Materials for PCR product purification, gel extraction, plasmid preparation, and Ni-NTA agarose were from Qiagen (Valencia, CA). The gene replacement vector, pKO3, was a generous gift from Dr. George M. Church (Harvard Medical School, Boston, MA). Asimicin was synthesized according to Ref. 38. The BCA protein assay kit and SuperSignal West Pico chemiluminescent substrate were from Pierce. Anti-His tag antibody was purchased from Abgent (San Diego, CA). All chemicals used were reagent grade.
Preparation of nuoE Knock-out and Mutant Strains Carrying the Gene for a His-tagged NuoE Subunit
The E. coli MC4100 strain was used to generate knock-out and site-specific mutations of nuoE employing the pKO3 system with the modification described previously by Kao et al. (17). Oligonucleotides used in this study are listed in supplemental Table S1. A DNA fragment, which includes the nuoE gene with its upstream and downstream 1-kb DNA segments, was amplified from E. coli DH5α by PCR. E. coli nuoE knock-out was constructed by replacement of the nuoE gene in the nuo operon encoding E. coli complex I with the nuoE fragment disrupted by spc (spectinomycin) gene with pKO3/nuoE(spc). A total of six histidine residues were sequentially introduced into the N terminus of NuoE by PCR using the sense and antisense primers in which the codes for polyhistidines were added (supplemental Table S1). In subsequent steps, the pKO3/(His)6 or 9 nuoE plasmid was used for the construction of His-tagged nuoE mutant strains. The introduction of polyhistidine sequences in the chromosome was verified by direct DNA sequencing.
Bacterial Growth and Membrane Vesicle Preparation
The E. coli membranes were prepared according to (17). In brief, the cells were grown in 4.8 liters (800 ml × six flasks) of Terrific Broth medium at 33 °C until A600 of ∼7 and then harvested at 5,800 × g for 20 min. The cells were resuspended at 10% (w/v) in a buffer containing 50 mm Bis-Tris (pH 6.0), 0.1 mm EDTA, 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride (PMSF), and 10% (w/v) glycerol. The cell suspensions were sonicated twice with a 1-s on/1-s off cycle for 60 s, passed once in a French press at 25,000 p.s.i., and centrifuged at 23,400 × g for 20 min. The supernatant was ultracentrifuged at 256,600 × g for 60 min. The pellet was resuspended in the buffer containing 50 mm Bis-Tris (pH 6.0 at 4 °C), 2 mm CaCl2, 10 mm 2-mercaptoethanol, and 10% glycerol, and a protease inhibitor mixture (Complete EDTA-free tablets, Roche Diagnostics) (buffer A) was added. The resulting membrane suspension was frozen in liquid nitrogen and stored at −80 °C until use.
Isolation of Complex I
All steps were carried out at 4 °C. The pellet (membrane fraction) was suspended at 5 mg of protein/ml in buffer A. After the addition of dodecyl-β-d-maltoside (DDM) at a final concentration of 1.2% (w/v), the sample was gently stirred for 30 min and centrifuged at 250,000 × g for 20 min. After this, 200 mm NaCl and 5 mm histidine (at the final concentrations) were added drop by drop to the supernatant. This was followed by the addition of Ni-NTA resins (Qiagen) equilibrated in 50 mm Bis-Tris, pH 6.0. The supernatant was incubated with gentle stirring for 1 h. Then, the sample was poured into an empty column. The gravity packed column was washed with 3–5 column volumes of buffer A containing 200 mm NaCl, 0.1% DDM, and 5–10 mm histidine, and the enzyme was eluted with buffer A containing 200 mm NaCl, 0.1% DDM, and 200 mm histidine. The brown-colored fractions were pooled and concentrated (Amicon-Ultra, molecular weight cut-off 100K; Millipore) to 2–8 mg of protein/ml. The concentrated complex I was immediately applied onto a desalting column (Econo-Pac 10DG, 10 ml; Bio-Rad), which had been equilibrated with buffer A containing 200 mm NaCl and 0.1% DDM, to remove histidine. The desalted enzyme fraction was quickly frozen in liquid nitrogen and stored at −80 °C until use.
Gel Electrophoresis and Western Blot Analysis
SDS-PAGE was performed according to Laemmli (39) and Schägger (40). The expression of the NuoE subunit was immunochemically determined using antibodies specific to NuoE. The assembly of E. coli complex I was evaluated by using Blue-Native PAGE according to Refs. 41 and 42. The position of the complex I band was confirmed by activity staining with nitroblue tetrazolium (16).
Quantification of Bound Quinones
The bound quinones in purified complex I were basically determined according to Shinzawa-Itoh et al. (43) except that we used ∼1 mg of the complex I/extraction. We also isolated ubiquinone 8 (UQ8) and menaquinone 8 (MK8) from E. coli membranes grown aerobically and semi-anaerobically and used them as standards. The 20-μl portion of each sample was applied to an HPLC system (LC-20AB, Shimadzu, Kyoto, Japan) equipped with a C18 column (Inertsil ODS3, 5 μm, 4.6 × 250 mm; GL science, Torrance, CA) equilibrated with a mobile phase consisting of ethanol/methanol/acetonitrile (4:3:3 v/v/v) at a flow rate of 1 ml/min at 50 °C. The elution was monitored on a UV-visible detector (SPD-20AV, Shimadzu, Kyoto, Japan) at 260 and 320 nm, and the content of UQ8 was estimated from the peak area by comparison with authentic compounds of known concentrations based on the molecular extinction, 14.9 mm−1 cm−1 at 275 nm reported (44).
Quantitative Analyses of FMN
The isolated complex I of 2 mg of protein was dissolved in 1 ml of 50 mm Bis-Tris buffer, pH 6.0, containing 0.1% DDM. The enzyme solution was treated with 5% trichloroacetic acid and homogenized with a Vortex mixer for 20 s and followed by heat treatment at 100 °C for 5 min. The denatured protein was removed by centrifugation. The FMN content was determined by the absorbance difference between 445 and 580 nm using the molecular extinction coefficient of FMN, Δϵ445–580 nm, of 11.1 mm−1 cm−1 in the presence of 5% trichloroacetic acid (43).
Activity Analyses
The NADH:K3Fe(CN)6, NADH:hexaamine ruthenium), and NADH:n-decylubiquinone activity was spectrophotometrically measured at 30 °C using a Cary 60 UV-visible spectrophotometer (Agilent, Santa Clara, CA). The buffers used were: (a) 50 mm MES buffer (pH 6.0) containing 50 mm NaCl, (b) 10 mm potassium phosphate buffer (pH 7.0) containing 1 mm EDTA, and (c) 25 mm HEPES-Bis-Tris propane (pH 7.5) containing 3 mm KCl. Reaction mixtures contained 150 μm NADH, 10 mm KCN, and 1 mm K3Fe(CN)6 and 350 μm hexaamine ruthenium or 25 μm n-decylubiquinone. Extinction coefficients of ϵ340 = 6.22 mm−1 cm−1 for NADH and ϵ420 = 1.00 mm−1 cm−1 for K3Fe(CN)6 were used for activity calculations. Reported values are the average of three measurements.
EPR Spectroscopy
EPR samples were prepared under strict anaerobic conditions. After the addition of 6 mm NADH or 10–20 mm neutralized sodium dithionite solution, each sample was incubated for 10 min, transferred to an EPR tube, and then frozen immediately. EPR spectra were recorded by a Bruker Elexsys E500 spectrometer at X-band (9.4 GHz) using an Oxford Instrument ESR900 helium flow cryostat. EPR spectra of the iron-sulfur clusters were simulated by SimFonia (Bruker). Power saturation data were analyzed by a computer fitting method as described previously (37, 45).
Other Analytical Procedures
Protein assay was routinely done by the methods of Lowry et al. (46) and/or Bradford (47) using bovine serum albumin as the standard. Iron content was determined by a colorimetric assay using ferrozine (48). Any variations from the procedures and other details are described in the figure legends.
RESULTS
Affinity Purification of E. coli Complex I
After the second round of PCR to extend three additional histidines in the N terminus of the NuoE subunit, we obtained the final plasmid containing (His)9-nuoE in addition to the originally designed (His)6-nuoE plasmid. The NuoE deletion strain was complemented by pKO3 plasmids containing either (His)6-nuoE or (His)9-nuoE, and we obtained both (His)6-nuoE and (His)9-nuoE mutant strains. Both mutant strains showed normal growth. Complex I activities in membrane fractions were similar to those in the wild type (data not shown). Western blotting using anti-NuoE antibody revealed that the size of the NuoE subunit was slightly increased as expected (Fig. 1A, left) and that the same band reacted with anti-His tag antibody (Fig. 1A, right). Because a longer polyhistidine tag increases binding affinity to the Ni-NTA resin and results in greater purity, the (His)9-nuoE strain was used for purification of complex I.
FIGURE 1.
Gel analysis of His-tagged complex I. A, immunoblotting analysis of E. coli membranes from wild type (lane 1) and the (His)9-nuoE mutant (lane 2). Membranes (20 μg/lane) were loaded onto a 12.5% Laemmli SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred onto nitrocellulose membranes, and Western blotting was carried out using the SuperSignal West Pico system. Antibodies specific to NuoE and His tag were used. B, SDS-PAGE of purified complex I. Tricine SDS-PAGE on a 10% gel was performed according to Ref 40. C, Blue-Native PAGE of purified complex I (CI). The arrows show the location of the fully assembled complex I and NuoEFG subcomplex. Blue-Native gels were stained with Coomassie Brilliant Blue (left) or incubated with 2.5 mg/ml nitro blue tetrazolium and 150 μm NADH for 1 h at 37 °C (right).
Cytoplasmic membranes were routinely prepared from ∼50 g of E. coli cells from 4.8 liters of culture. About 70% of membrane proteins were extracted with 1.2% DDM (lipid:protein = 2.4:1). The concentration of histidine in the binding and washing buffer for the Ni-NTA resin was critical to obtain highly pure complex I. We found that the presence of 5 mm histidine in the buffer drastically reduced contamination to almost zero. The specific activity became 25 times higher at the affinity column step (Table 1). Overall, 10% of total NADH:ferricyanide activity was recovered. NADH:ferricyanide and NADH:n-decylubiquinone activities of the final preparation were 219 ± 9.7 and 39.2 ± 8 μmol/min/mg, respectively, by using HEPES-Bis-Tris propane, pH 7.5. We found that NADH:ferricyanide activities varied depending on the buffers used (supplemental Table S2), although NADH:ferricyanide activities are not pH-dependent (49).
TABLE 1.
Isolation of E. coli complex I from strain MC4100/His9 (NuoE)
| Preparation | Volume | Protein | NADH:ferricyanide activitya |
Yield | |
|---|---|---|---|---|---|
| Total activity | Specific activity | ||||
| ml | mg | μmol/min | μmol/min × mg | % | |
| Membranes | 35 | 1100 | 7056 | 6.41 | 100 |
| Extract | 225 | 909 | 4860 | 5.35 | 68.9 |
| Ni-NTA | 7.5 | 3.3 | 438 | 134.0 | 6.2 |
| MWCOb 100 kDa | 1.15 | 3.2 | 707 | 219.0 | 10.0 |
a HEPES-BTP 25 mm (pH 7.5) buffer containing 3 mm KCl was used.
b MWCO, molecular weight cut-off.
The SDS-PAGE pattern of purified complex I demonstrated the presence of all complex I subunits (Fig. 1B). No impurities were detected. There was almost no ∼85-kDa band, which has been frequently detected in purified complex I and identified as a proteolytically cleaved fragment of NuoG (50). We further verified the assembly of the His-tagged complex I assembly by Blue-Native PAGE with NADH dehydrogenase activity staining. As shown in Fig. 1C, the one major band was detected around the molecular mass of ∼570 kDa, which is close to the estimated molecular mass, 535 kDa, based on the amino acid sequences of 13 subunits NuoA-N in E. coli complex I. The more densely stained band around 170 kDa corresponds to the NADH dehydrogenase complex. Due to the low concentration of DDM (0.1%) in the purified sample, some portion of purified complex I disintegrated during electrophoresis. Clearly, the polyhistidine insertion in NuoE did not affect the subunit assembly of complex I.
The purified complex I contained 0.94 ± 0.1 of FMN and 29.0 ± 0.37 of iron/1 mol of CI, respectively. The complex I preparation exhibited absorption spectra as shown in Fig. 2A. It has a sharp peak at 274 nm and two broad shoulders at 410 and 560 nm. This feature was consistent with the previous study (51). No contaminant hemeproteins were detectable. The apparent molecular extinction coefficients of peaks at 274 nm (or at 280 nm) and 410 nm were 495 (472) and 50.3 mm−1 cm−1, respectively. The broad shoulders from 400–600 nm were quenched by dithionite (Fig. 2A, inset).
FIGURE 2.
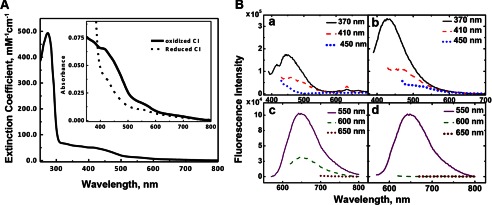
A, UV-visible absorption spectra of purified E. coli complex I. Complex I was dissolved in 50 mm Bis-Tris (pH 6.0), 2 mm CaCl2, 10% glycerol, and 0.1% DDM at 0.73 mg/ml. Inset, the enlarged absorption spectra from 350 to 800 nm. Thick solid line, the oxidized form; dotted line, the reduced form by the addition of 10 mm sodium dithionite. B, fluorescence emission spectra of purified E. coli complex I at various excitation wavelengths (370, 410, 450, 550, 600, and 650 nm). Fluorescence emission spectra were taken from the oxidized enzyme (panels a and c) and reduced enzyme with 10 mm dithionite (panels b and d).
Fluorescence spectra of the purified complex I were also measured. We detected a weak, but substantial fluorescence emission, maximal at ∼445 nm in the oxidized state (Fig. 2B, panel a). The signal was doubled and shifted to ∼425 nm in the reduced state (Fig. 2B, panel b). Their excitation spectra suggest that these emission signals originate from the flavin chromophore. Interestingly, we found an additional fluorescent emission peak at ∼650 nm regardless of the redox state of the protein (Fig. 2B, panels c and d), which originates from an excitation peak at 280 nm and 560 nm (data not shown). This feature somewhat resembles the luminescent properties of NADH dehydrogenase-2 (NDH-2) from E. coli, which was predicted to bind a copper (52). This possibly suggests that there might be a metal binding to complex I.
The ubiquinone content of complex I purified from aerobically grown E. coli cells was determined by cyclohexane extraction and HPLC measurements (Fig. 3). The amount of ubiquinone was found to be 1.99 ± 0.07 mol of ubiquinone/1 mol of complex I. The result opposed previous studies in which only one ubiquinone molecule is bound to E. coli (53) and bovine (43) complex I. It is known that UQ8 and MK8 are major quinone components in E. coli (54). In fact, our complex I preparations isolated from cells grown in semi-anaerobic conditions contain a significant amount of MK8 as well as UQ8 (data not shown).
FIGURE 3.
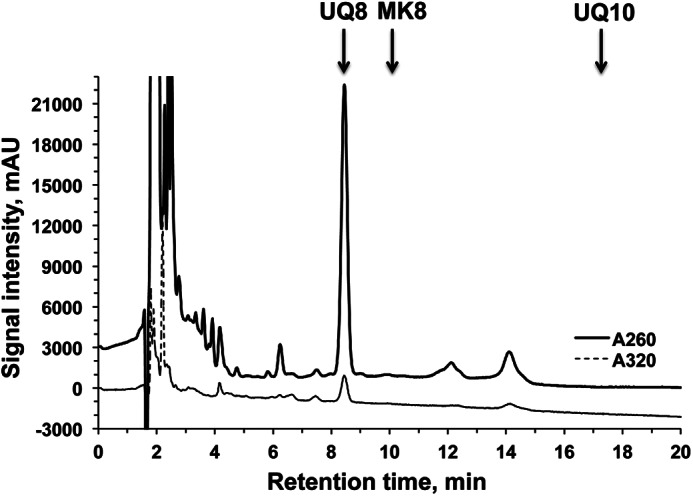
HPLC analysis of quinones extracted from purified complex I using cyclohexane. The chromatograms were shown at 260 and 320 nm. Arrows indicate the elution times for standards UQ8, MK8, and UQ10. mAU, milliabsorbance units.
EPR Analysis
The iron-sulfur clusters of the preparations were characterized by X-band EPR spectroscopy. EPR signals from binuclear clusters N1a and N1b were observed at 40 K and 2 mW (Fig. 4A). Their signal intensities were almost the same in both NADH-reduced and dithionite-reduced samples (Fig. 4A, spectra a and b). The EPR signals of the tetranuclear iron-sulfur clusters N2, N3, and N4 were present in the spectra of both NADH-reduced and dithionite-reduced samples recorded at 13 K and 5 mW microwave power in addition to the signals of the binuclear clusters (Fig. 4B). No obvious difference was detectable concerning the gz values and line width of the signals from those clusters. The signal intensities at gz = 2.05 and 2.03 and in the gy = ∼1.94 region, however, were much larger in the dithionite-reduced sample than those in the NADH-reduced sample. This difference between NADH- and dithionite-reduced seemed to be mostly caused by EPR signals from cluster N7, which cannot be reduced with NADH but can be reduced with dithionite due to its long distance (20.5 Å) from the main electron pathway (55). Therefore, previously assigned signals were all present at the expected positions in our complex I preparation. Their relative signal intensities were similar to those in published literature (56).
FIGURE 4.
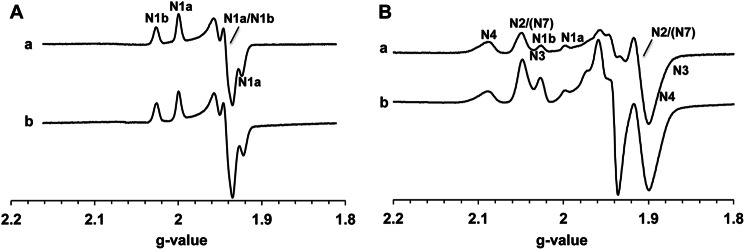
EPR spectra of the isolated complex I. The samples (8.28 mg/ml) were anaerobically reduced with 6 mm NADH (spectrum a) or 20 mm dithionite (spectrum b). The EPR spectra were recorded at 40 K and 2 mW (A) or 13 K and 5 mW (B). The position of the EPR signals arising from each iron-sulfur cluster is indicated. Other EPR conditions were: microwave frequency, 9.45 GHz; modulation frequency, 100 kHz; modulation amplitude, 6 G; time constant, 82 ms.
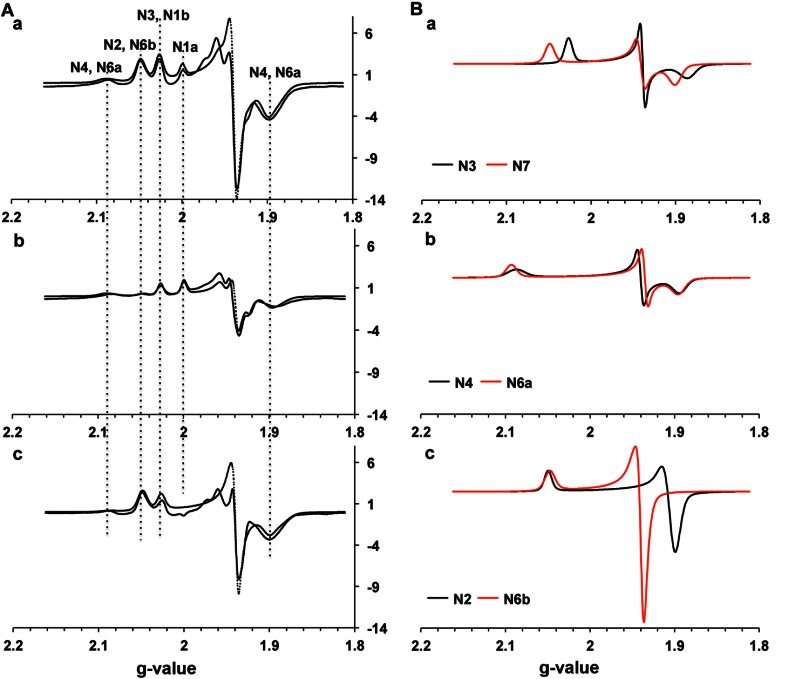
Clusters N6a and N6b are historically believed to be EPR silent signals. The recent mutational study by the Yagi group suggest that the gz,x = 2.08, 1.88 signals are arising from cluster N6a (35). Therefore, we carried out more detailed EPR power saturation analysis at different temperatures. For the first time, we found that there were overlapping EPR signals at ∼2.09 in E. coli complex I as shown in Fig. 5. The g = 2.099 signal showed much higher intensity at 204 mW than at 1.2 mW (Fig. 5, spectra a and b), indicating that it has faster spin relaxation, whereas the g = 2.086 signal showed the opposite; its intensity is larger at 1.2 mW than at 204 mW (Fig. 5, spectra c and d). There are two peaks in the difference spectrum at 81 mW (dithionite-reduced minus NADH-reduced, Fig. 5, spectrum e). These data strongly suggest that the cluster N6a signal does exist in the spectra of purified complex I, in addition to cluster N4. Previously, Yano et al. (57) showed the EPR signals of clusters N6a and N6b in the overexpressed Paracoccus denitrificans Nqo9 (an E. coli NuoI homologue) subunit: an outside-pair gz,x = 2.08 and 1.89 and an inside-pair gz,x = 2.05 and 1.92. Because the outside-pair signals showed much faster spin relaxation (P½ = 342 mW) than the inside-pair signals (P½ = 8 mW) at 14 K, the g = 2.099 was assigned to cluster N6a, and the g = 2.086 signal was assigned to cluster N4 (Fig. 5).
FIGURE 5.
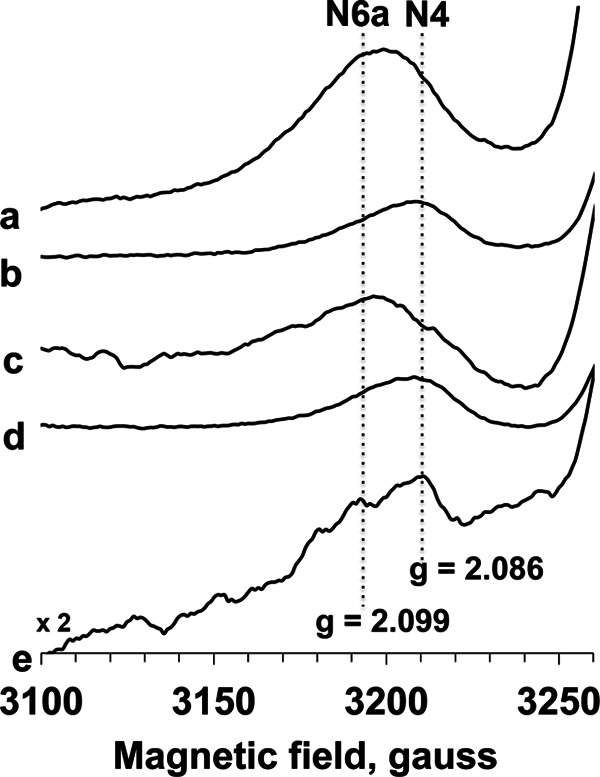
EPR spectra of the gz region of cluster N4 and N6a signals in E. coli complex I. The samples were anaerobically reduced with 20 mm dithionite (spectra a and b) or 6 mm NADH (spectra c and d). The EPR spectra were recorded at 10 K and 204 mW (spectra a and c) or 1.2 mW (spectra b and d). The difference spectra (dithionite-reduce minus NADH-reduced) were taken at 10 K and 81 mW (spectrum e). Other EPR conditions are the same as in Fig. 4.
The gz signal of the inside-pair gz,x = 2.05 and 1.92 signals detected in the overexpressed P. denitrificans Nqo9 arising from cluster N6b is completely overlapped with the gz signal of cluster N2. In the search for cluster N6b signals in our complex I samples, we further obtained difference spectra at different temperatures and different microwave powers and analyzed the data by using simulation techniques. We observed that the reduction levels of iron-sulfur clusters in complex I were the same or higher in dithionite-reduced spectra than in NADH-reduced spectra. We decided to use EPR data at 20 K and 5 mW (Fig. 6), minimizing the contribution from other tetranuclear clusters in difference spectra (dithionite-reduced minus NADH-reduced). Based on the long accepted fact that cluster N2 is fully reduced with NADH (13, 58), the dithionite-reduced samples, the NADH-reduced samples, and their difference spectra at 20 K and 5 mW were individually simulated using the g-value parameters for EPR signals of each iron-sulfur cluster (supplemental Table S3). As shown in Fig. 6A, panel c, the dithionite-reduced minus NADH-reduced difference spectrum revealed additional axial type EPR signals at g = 2.05 and 1.93, which cannot be simulated using parameters for clusters N3 (gz,y,x = 2.03, 1.94, 1.88, Fig. 6B, panel a), N4 (gz,y,x = 2.09, 1.94, 1.89, Fig. 6B, panel b), and N7 (gz,y,x = 2.05, 1.94, 1.90, Fig. 6B, panel a). This strongly suggested the presence of the cluster N6b signals that can only be reduced with dithionite under strict anaerobic conditions. This nature is completely opposite to the nature of cluster N2 signals, which can completely be reduced with NADH even under aerobic conditions. However, both N2 and N6b signals are axial and detectable at higher temperatures. The spectrum (Fig. 6B, panel c, red line) shows the simulation of the axial-type EPR signal arising from cluster N6b, overlapped with the gz value of the cluster N2 signal. The dotted line in Fig. 6A, panel c, shows the sum of simulated spectra using the following spin contributions of individual clusters: N3:N4:N6b:N7 = 0.77:0.19:0.31: 1.
FIGURE 6.
A, fitting the experimental EPR spectra of complex I reduced by dithionite (panel a) or NADH (panel b) and their difference (dithionite-reduced minus NADH-reduced) spectra (panel c) with simulated EPR signals of individual iron-sulfur clusters. The spectra were recorded at 20 K and 5 mW and are displayed at the same scale. The dotted lines show the sum of the simulated spectra of each iron-sulfur clusters. B, simulated EPR signals of individual tetranuclear clusters. The spectra are normalized to give the same spin quantity (panels a–c).
Semiquinone Signals in Purified E. coli Complex I
SQ signals have never been detected in purified E. coli complex I from other research laboratories. However, we detected a small but significant SQ signal at 20 K (Fig. 6A, panel c, in the g = 2.00 region). We examined the power saturation profiles of this g = 2.005, SQ signal at 150 K. Unfortunately, even at 150 K, the relatively large gz = 2.00 signal from cluster N1a was still overlapped with the SQ signal (Fig. 7, spectrum a). Thus, we subtracted the dithionite-reduced spectra from NADH-reduced spectra to isolate the SQ signal. This was feasible because cluster N1a was completely reduced by either NADH or dithionite, whereas the SQ signal disappeared in the dithionite-reduced spectra (Fig. 7, spectrum c). Finally, we successfully isolated the SQ signal in E. coli complex I (Fig. 7, top right). The peak-to-peak line width (ΔHpp) was ∼12 G, indicating its neutral semiquinone form (QH•) (59). It is known that protonation increases spectral line width because of an asymmetric perturbation of the spin density on the quinone ring. Based on power saturation analysis, more than 90% of the SQ signal originated from the single entity with P½ (half-saturation power level) = 1.85 mW. There was no flavosemiquinone component that does not saturate above 10 mW and has a broader ΔHpp, typically >15 G (60, 61). It was reported that if complex I was chemically reduced in the potentiometric titration, the ΔHpp value of flavosemiquinone in complex I was around 24 G (60).
FIGURE 7.
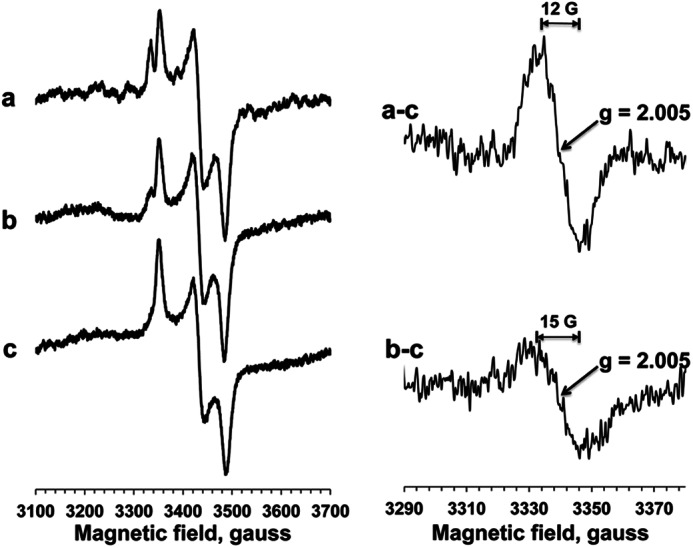
EPR spectra of SQ signals in isolated complex I. Left, EPR spectra were recorded at 150 K and 1.2 mW. Complex I (15 μm) was reduced with NADH in the absence (spectrum a) or presence (spectrum b) of asimicin, a potent E. coli complex I inhibitor, or reduced with dithionite (spectrum c). Right, the difference spectra of NADH-reduced minus dithionite-reduced spectra in the absence (spectrum a-c) or presence (spectrum b-c) of asimicin.
Furthermore, the SQ signal was decreased by 60% in the presence of asimicin (100 μm) (20), a potent inhibitor of E. coli complex I (Fig. 7, spectrum b). This phenomenon can be seen more clearly in the difference spectrum of the NADH-reduced spectra minus the dithionite-reduced spectra (Fig. 7, bottom right). Our data strongly suggest that this SQ signal is likely to be involved in redox-coupled proton-pumping mechanism of complex I.
DISCUSSION
Inserting a polyhistidine coding sequence into the N terminus of the NuoE subunit of complex I allowed for highly efficient and rapid affinity purification of this enzyme, overcoming its low abundance in E. coli membranes. We obtained fully assembled, highly pure, and active complex I, in which all cofactors FMN, iron-sulfur clusters, and bound quinones are present in stoichiometric amounts. Using these preparations, we successfully detected SQ signals for the first time in E. coli complex I. In addition, we detected two additional EPR signals arising from clusters N6a and N6b, which have been believed to be undetectable by EPR.
UQ Binding Sites and Semiquinone Signals
It is thought that complex I contains two quinone binding sites (62). One UQ site is located in the vicinity of cluster N2, whereas the second UQ site is in the hydrophobic membrane relatively far from cluster N2 where UQ is easily exchanged in the quinone pool. In fact, two protein-associated SQ species in complex I have been characterized using tightly coupled bovine heart submitochondrial particles. They are distinguished as SQNf (fast) and SQNS (slow), based on their spin relaxation properties. Both signals are sensitive to complex I inhibitors, whereas only SQNf signal is uncoupler-sensitive. Using a powerful program to analyze exchange and dipolar interactions, the center to center distance between and SQNf has been estimated to be 12 Å (63), whereas the SQNS is located >30 Å away from cluster N2, suggesting that SQNs is not the direct electron acceptor from cluster N2 (64). Because of the high stability constant of the SQNs (2.0), it has been suggested that SQNs functions as a converter between n = 1 and n = 2 electron transfer steps (64).
Because our SQ signal was detected under no proton motive force (Δp), we believe that this SQ signal corresponds to the slow relaxing SQNs in bovine complex I. The SQNs signal in isolated bovine complex I after aerobic reduction with NADH was detected in an anionic form at pH 7.8 with ΔHpp = 6.1 G at 173 K, and its spin relaxation rate (P½) was 0.57 mW (64). This value is reasonably comparable with the P½ data (1.85 mW) of our SQ signal at 150 K. Interestingly, the SQ signal in our E. coli complex I was detected in a neutral semiquinone form (QH•) at pH 6 with ΔHpp = 12 G (Fig. 7, top right). On the other hand, in bovine complex I, the SQNf and SQNS signals were both reported to be anionic at pH 8.0. In the recent model of the proton translocation mechanism proposed by Treberg and Brand (65), protonations and deprotonations of Q at some steps were added to incorporate their observation of ΔpH dependence of superoxide formation from SQ. Although it is not clear whether QH• or Q•− as described in their model actually corresponds to SQNf or SQNS, the experimental fact that the SQ signal detected in our E. coli complex I was neutral at pH 6.0 fits this prediction. More detailed characterization of this SQ signal including pH dependence and the redox mid-potential (Em,7) for the Q/SQ couple is underway.
Another interesting fact was that we did not observe any flavosemiquinone component when isolated complex I was reduced with far excess amount (400–1000 times) of NADH under anaerobic conditions. In the presence of the potent E. coli complex I inhibitor asimicin, the SQ signal was decreased by 60%. The remaining asimicin-insensitive SQ signal showed the somewhat larger ΔHpp value of ∼15 G, but this signal was saturated at higher microwave power. Additionally, its P½ was 1.20 mW, which is even slightly smaller than 1.84 mW, the P½ value of the asimicin-sensitive SQ signal. These results offer a large contrast to the results for bovine complex I, in which the flavosemiquinone signal is enhanced when no catalytic oxidation of NADH occurs or NADH oxidation is inhibited by complex I inhibitors (61). This difference may partly explain why bovine complex I produces 95% superoxide, whereas E. coli complex I produces ∼20% superoxide and 80% hydrogen peroxide, when complex I reduces O2 (66).
It remains controversial as to whether the two semiquinone species SQNf and SQNS reflect the same semiquinone intermediate bound to the well characterized ubiquinone binding pocket in the vicinity of cluster N2 but in different conformational states of the enzyme or whether they are distinct semiquinone species localized in two different quinone binding sites: the pocket in the vicinity of cluster N2 and the hydrophobic membrane pocket relatively far from cluster N2. In either case, the sum of the spin concentrations of the two semiquinone species was never found to exceed one per complex I protein. In addition, the Sazanov group (12) strongly argued against the existence of the second Q binding site based on the recent crystal structure of the entire T. thermophilus complex I. Therefore, it might be very interesting to investigate the nature of SQNs and its relationship to SQNf in our complex I preparations, which contain 1.99 ± 0.07 mol of ubiquinone/1 mol of complex I.
Cluster N6a and N6b Signals
The gz = 2.09 signal of cluster N6a was previously detected based on mutation experiments, whereas other changes in the gz = 2.09 region were not observed in their partially purified E. coli complex I samples (35). However, using highly pure and concentrated samples, we were able to detect the heterogeneity of the gz = 2.09 signals arising from both clusters N4 and N6b and isolate their peaks for the first time (Fig. 5). The similar heterogeneous behaviors showing different temperature and power dependence have also been observed in the g = 2.10 and 1.88 regions of EPR signals from purified bovine heart complex I (Fig. 4 in Ref. 67). Therefore, our results strongly suggest that two overlapping EPR signals arising from clusters N4 and N6a in the gz = 2.09–2.10 exist in both E. coli and bovine complex I. Based on the temperature dependence and spin relaxation features of cluster N6a in the overexpressed subunit, we assigned the outside g = 2.099 signal to cluster N6a and the g = 2.086 signal to cluster N4. In 1975, Ohnishi (31) potentiometrically distinguished two signals around g = 2.10–2.11 using pigeon heart submitochondrial particles. The g = 2.10 and 2.11 signals exhibited different Em,7.2 values, −240 and −410 mV, respectively. This result could explain our observation that the outside g = 2.099 (cluster N6a) signal was not efficiently reduced with NADH compared with dithionite (Fig. 5).
To isolate cluster N6b signals from the entire EPR spectra, we subtracted NADH-reduced from dithionite-reduced spectra at 20 K and 5 mW and simulated using parameters for each iron-sulfur cluster. At 20 K, there should be no signal from the fast relaxing N6a cluster. The contribution of EPR signals from clusters N1a, N1b, and N2 can mostly be neglected because clusters N1a, N1b, and N2 are reduced to the same extent with NADH or dithionite. Thus, the resulting difference spectrum is the sum of EPR signals arising from clusters N3, N4, N6b, and N7. We successfully assigned the axial signal (gz,y,x = 2.05, 1.94, 1.94) to cluster N6b (Fig. 6B, panel c). The recent mutational analyses by the Yagi group (35) also suggest that the signal of gz = 2.05 is also arising from cluster N6b.
It is noteworthy that both clusters N6a and N6b in our highly pure E. coli complex I preparations are efficiently reduced with dithionite but not with NADH under strict anaerobic conditions. In particular, cluster N6b is not reduced with NADH. This result is consistent with the results by Mössbauer spectroscopy (58). Besides the problem of extensively overlapped EPR signals in complex I, another reason why the EPR signals arising from clusters N6a and N6b have not been observed might be that most EPR measurements have been done with NADH-reduced samples.
The proton-pumping mechanism of complex I is still largely unknown, although more detailed crystal structures of the entire complex I from T. thermophilus have recently been determined at 3.3 Å (12). In this study, we have shown that our highly pure and active complex I was suitable for sophisticated biochemical and biophysical analyses including EPR simulation. Combined with available structural information, our highly pure complex I will facilitate the study of a redox-driven proton-pumping mechanism of complex I in the near future.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM097409 (to E. N.-O.). This work was also supported by American Heart Association Grant 11SDG5560001 (to E. N.-O.).

This article contains supplemental Tables S1–S3.
- UQ
- ubiquinone
- Q
- quinone
- SQ
- semiquinone
- MK
- menaquinone
- Ni-NTA
- nickel-nitrilotriacetic acid
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- mW
- milliwatt(s)
- DDM
- dodecyl-β-d-maltoside
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Yagi T., Matsuno-Yagi A. (2003) The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry 42, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 2. Sazanov L. A. (2007) Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46, 2275–2288 [DOI] [PubMed] [Google Scholar]
- 3. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 4. Hinkle P. C., Kumar M. A., Resetar A., Harris D. L. (1991) Mechanistic stoichiometry of mitochondrial oxidative phosphorylation. Biochemistry 30, 3576–3582 [DOI] [PubMed] [Google Scholar]
- 5. DiMauro S., Schon E. A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- 6. Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 7. Chomyn A., Cleeter M. W., Ragan C. I., Riley M., Doolittle R. F., Attardi G. (1986) URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science 234, 614–618 [DOI] [PubMed] [Google Scholar]
- 8. Chomyn A., Mariottini P., Cleeter M. W., Ragan C. I., Matsuno-Yagi A., Hatefi Y., Doolittle R. F., Attardi G. (1985) Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature 314, 592–597 [DOI] [PubMed] [Google Scholar]
- 9. Yagi T., Yano T., Di Bernardo S., Matsuno-Yagi A. (1998) Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364, 125–133 [DOI] [PubMed] [Google Scholar]
- 10. Yip C. Y., Harbour M. E., Jayawardena K., Fearnley I. M., Sazanov L. A. (2011) Evolution of respiratory complex I: ”supernumerary“ subunits are present in the α-proteobacterial enzyme. J. Biol. Chem. 286, 5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 12. Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A. (2013) Crystal structure of the entire respiratory complex I. Nature 494, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohnishi T. (1998) Iron-sulfur clusters/semiquinones in complex I. Biochim. Biophys. Acta 1364, 186–206 [DOI] [PubMed] [Google Scholar]
- 14. Ohnishi T., Nakamaru-Ogiso E. (2008) Were there any “misassignments” among iron-sulfur clusters N4, N5, and N6b in NADH-quinone oxidoreductase (complex I)? Biochim. Biophys. Acta 1777, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sazanov L. A., Hinchliffe P. (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 16. Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2007) Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 282, 36914–36922 [DOI] [PubMed] [Google Scholar]
- 17. Kao M. C., Di Bernardo S., Perego M., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2004) Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 279, 32360–32366 [DOI] [PubMed] [Google Scholar]
- 18. Kao M. C., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 44, 9545–9554 [DOI] [PubMed] [Google Scholar]
- 19. Kao M. C., Di Bernardo S., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoJ (ND6) of the NADH-quinone oxidoreductase from Escherichia coli by chromosomal DNA manipulation. Biochemistry 44, 3562–3571 [DOI] [PubMed] [Google Scholar]
- 20. Nakamaru-Ogiso E., Kao M. C., Chen H., Sinha S. C., Yagi T., Ohnishi T. (2010) The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J. Biol. Chem. 285, 39070–39078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murai M., Ishihara A., Nishioka T., Yagi T., Miyoshi H. (2007) The ND1 subunit constructs the inhibitor binding domain in bovine heart mitochondrial complex I. Biochemistry 46, 6409–6416 [DOI] [PubMed] [Google Scholar]
- 22. Nakamaru-Ogiso E., Sakamoto K., Matsuno-Yagi A., Miyoshi H., Yagi T. (2003) The ND5 subunit was labeled by a photoaffinity analogue of fenpyroximate in bovine mitochondrial complex I. Biochemistry 42, 746–754 [DOI] [PubMed] [Google Scholar]
- 23. Yagi T., Hatefi Y. (1988) Identification of the dicyclohexylcarbodiimide-binding subunit of NADH-ubiquinone oxidoreductase (Complex I). J. Biol. Chem. 263, 16150–16155 [PubMed] [Google Scholar]
- 24. Nakamaru-Ogiso E., Han H., Matsuno-Yagi A., Keinan E., Sinha S. C., Yagi T., Ohnishi T. (2010) The ND2 subunit is labeled by a photoaffinity analogue of asimicin, a potent complex I inhibitor. FEBS Lett. 584, 883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tocilescu M. A., Zickermann V., Zwicker K., Brandt U. (2010) Quinone binding and reduction by respiratory complex I. Biochim. Biophys. Acta 1797, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 26. Shiraishi Y., Murai M., Sakiyama N., Ifuku K., Miyoshi H. (2012) Fenpyroximate binds to the interface between PSST and 49 kDa subunits in mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry 51, 1953–1963 [DOI] [PubMed] [Google Scholar]
- 27. Ohnishi T., Sled V. D., Yano T., Yagi T., Burbaev D. S., Vinogradov A. D. (1998) Structure-function studies of iron-sulfur clusters and semiquinones in the NADH-Q oxidoreductase segment of the respiratory chain. Biochim. Biophys. Acta 1365, 301–308 [DOI] [PubMed] [Google Scholar]
- 28. Nakamaru-Ogiso E., Matsuno-Yagi A., Yoshikawa S., Yagi T., Ohnishi T. (2008) Iron-sulfur cluster N5 is coordinated by an HXXXCXXCXXXXXC motif in the NuoG subunit of Escherichia coli NADH:quinone oxidoreductase (complex I). J. Biol. Chem. 283, 25979–25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakamaru-Ogiso E., Yano T., Ohnishi T., Yagi T. (2002) Characterization of the iron-sulfur cluster coordinated by a cysteine cluster motif (CXXCXXXCX27C) in the Nqo3 subunit in the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Thermus thermophilus HB-8. J. Biol. Chem. 277, 1680–1688 [DOI] [PubMed] [Google Scholar]
- 30. Nakamaru-Ogiso E., Yano T., Yagi T., Ohnishi T. (2005) Characterization of the iron-sulfur cluster N7 (N1c) in the subunit NuoG of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 280, 301–307 [DOI] [PubMed] [Google Scholar]
- 31. Ohnishi T. (1975) Thermodynamic and EPR characterization of iron-sulfur centers in the NADH-ubiquinone segment of the mitochondrial respiratory chain in pigeon heart. Biochim. Biophys. Acta 387, 475–490 [DOI] [PubMed] [Google Scholar]
- 32. Yakovlev G., Reda T., Hirst J. (2007) Reevaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 104, 12720–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velazquez I., Nakamaru-Ogiso E., Yano T., Ohnishi T., Yagi T. (2005) Amino acid residues associated with cluster N3 in the NuoF subunit of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. FEBS Lett. 579, 3164–3168 [DOI] [PubMed] [Google Scholar]
- 34. Uhlmann M., Friedrich T. (2005) EPR signals assigned to Fe/S cluster N1c of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) derive from cluster N1a. Biochemistry 44, 1653–1658 [DOI] [PubMed] [Google Scholar]
- 35. Sinha P. K., Nakamaru-Ogiso E., Torres-Bacete J., Sato M., Castro-Guerrero N., Ohnishi T., Matsuno-Yagi A., Yagi T. (2012) Electron transfer in subunit NuoI (TYKY) of Escherichia coli NADH:quinone oxidoreductase (NDH-1). J. Biol. Chem. 287, 17363–17373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magnitsky S., Toulokhonova L., Yano T., Sled V. D., Hägerhäll C., Grivennikova V. G., Burbaev D. S., Vinogradov A. D., Ohnishi T. (2002) EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH-ubiquinone oxidoreductase (complex I) in situ. J. Bioenerg. Biomembr. 34, 193–208 [DOI] [PubMed] [Google Scholar]
- 37. Vinogradov A. D., Sled V. D., Burbaev D. S., Grivennikova V. G., Moroz I. A., Ohnishi T. (1995) Energy-dependent Complex I-associated ubisemiquinones in submitochondrial particles. FEBS Lett. 370, 83–87 [DOI] [PubMed] [Google Scholar]
- 38. Han H., Sinha M. K., D'Souza L. J., Keinan E., Sinha S. C. (2004) Total synthesis of 34-hydroxyasimicin and its photoactive derivative for affinity labeling of the mitochondrial complex I. Chemistry 10, 2149–2158 [DOI] [PubMed] [Google Scholar]
- 39. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 41. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 42. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 43. Shinzawa-Itoh K., Seiyama J., Terada H., Nakatsubo R., Naoki K., Nakashima Y., Yoshikawa S. (2010) Bovine heart NADH-ubiquinone oxidoreductase contains one molecule of ubiquinone with ten isoprene units as one of the cofactors. Biochemistry 49, 487–492 [DOI] [PubMed] [Google Scholar]
- 44. Crane F. L., Barr R. (1971) Determination of ubiquinones. Methods Enzymol. 18 Part C, 137–165 [Google Scholar]
- 45. Rupp H., Rao K. K., Hall D. O., Cammack R. (1978) Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim. Biophys. Acta 537, 255–260 [DOI] [PubMed] [Google Scholar]
- 46. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 47. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 48. Carter P. (1971) Spectrophotometric determination of serum iron at the submicrogram level with a new reagent (ferrozine). Anal. Biochem. 40, 450–458 [DOI] [PubMed] [Google Scholar]
- 49. Zickermann V., Kurki S., Kervinen M., Hassinen I., Finel M. (2000) The NADH oxidation domain of complex I: do bacterial and mitochondrial enzymes catalyze ferricyanide reduction similarly? Biochim. Biophys. Acta 1459, 61–68 [DOI] [PubMed] [Google Scholar]
- 50. Mamedova A. A., Holt P. J., Carroll J., Sazanov L. A. (2004) Substrate-induced conformational change in bacterial complex I. J. Biol. Chem. 279, 23830–23836 [DOI] [PubMed] [Google Scholar]
- 51. Braun M., Bungert S., Friedrich T. (1998) Characterization of the overproduced NADH dehydrogenase fragment of the NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochemistry 37, 1861–1867 [DOI] [PubMed] [Google Scholar]
- 52. Rapisarda V. A., Chehín R. N., De Las Rivas J., Rodríguez-Montelongo L., Farías R. N., Massa E. M. (2002) Evidence for Cu(I)-thiolate ligation and prediction of a putative copper-binding site in the Escherichia coli NADH dehydrogenase-2. Arch Biochem. Biophys 405, 87–94 [DOI] [PubMed] [Google Scholar]
- 53. Verkhovsky M., Bloch D. A., Verkhovskaya M. (2012) Tightly-bound ubiquinone in the Escherichia coli respiratory complex I. Biochim. Biophys. Acta 1817, 1550–1556 [DOI] [PubMed] [Google Scholar]
- 54. Mustafa G., Migita C. T., Ishikawa Y., Kobayashi K., Tagawa S., Yamada M. (2008) Menaquinone as well as ubiquinone as a bound quinone crucial for catalytic activity and intramolecular electron transfer in Escherichia coli membrane-bound glucose dehydrogenase. J. Biol. Chem. 283, 28169–28175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pohl T., Bauer T., Dörner K., Stolpe S., Sell P., Zocher G., Friedrich T. (2007) Iron-sulfur cluster N7 of the NADH:ubiquinone oxidoreductase (complex I) is essential for stability but not involved in electron transfer. Biochemistry 46, 6588–6596 [DOI] [PubMed] [Google Scholar]
- 56. Pohl T., Uhlmann M., Kaufenstein M., Friedrich T. (2007) Lambda Red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I). Biochemistry 46, 10694–10702 [DOI] [PubMed] [Google Scholar]
- 57. Yano T., Magnitsky S., Sled' V. D., Ohnishi T., Yagi T. (1999) Characterization of the putative 2×[4Fe-4S]-binding NQO9 subunit of the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Paracoccus denitrificans: expression, reconstitution, and EPR characterization. J. Biol. Chem. 274, 28598–28605 [DOI] [PubMed] [Google Scholar]
- 58. Bridges H. R., Bill E., Hirst J. (2012) Mössbauer spectroscopy on respiratory complex I: the iron-sulfur cluster ensemble in the NADH-reduced enzyme is partially oxidized. Biochemistry 51, 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bowyer J. R., Ohnishi T. (1985) EPR spedtrostudy in the study of ubisemiquinones in redox chains. in Coenxyme Q (Lenaz G., ed) pp. 409–432, John Wiley & Sons, New York [Google Scholar]
- 60. Sled V. D., Rudnitzky N. I., Hatefi Y., Ohnishi T. (1994) Thermodynamic analysis of flavin in mitochondrial NADH:ubiquinone oxidoreductase (complex I). Biochemistry 33, 10069–10075 [DOI] [PubMed] [Google Scholar]
- 61. Ohnishi S. T., Shinzawa-Itoh K., Ohta K., Yoshikawa S., Ohnishi T. (2010) New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): the significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochim. Biophys. Acta 1797, 1901–1909 [DOI] [PubMed] [Google Scholar]
- 62. Ohnishi S. T., Salerno J. C., Ohnishi T. (2010) Possible roles of two quinone molecules in direct and indirect proton pumps of bovine heart NADH-quinone oxidoreductase (complex I). Biochim. Biophys. Acta 1797, 1891–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yano T., Dunham W. R., Ohnishi T. (2005) Characterization of the ΔμH+-sensitive ubisemiquinone species (SQNf) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochemistry 44, 1744–1754 [DOI] [PubMed] [Google Scholar]
- 64. Ohnishi T., Johnson J. E., Jr., Yano T., Lobrutto R., Widger W. R. (2005) Thermodynamic and EPR studies of slowly relaxing ubisemiquinone species in the isolated bovine heart complex I. FEBS Lett. 579, 500–506 [DOI] [PubMed] [Google Scholar]
- 65. Treberg J. R., Brand M. D. (2011) A model of the proton translocation mechanism of complex I. J. Biol. Chem. 286, 17579–17584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Esterházy D., King M. S., Yakovlev G., Hirst J. (2008) Production of reactive oxygen species by complex I (NADH:ubiquinone oxidoreductase) from Escherichia coli and comparison to the enzyme from mitochondria. Biochemistry 47, 3964–3971 [DOI] [PubMed] [Google Scholar]
- 67. Nakamaru-Ogiso E. (2012) Iron-sulfur clusters in complex I. in A structural perspective on respiratory complex I (Sazanov L., ed) pp. 61–79, Springer, London [Google Scholar]