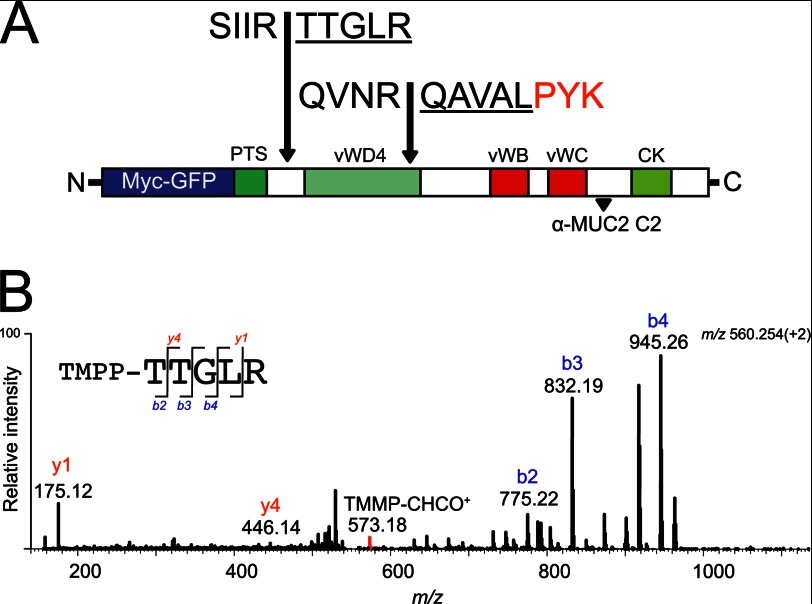
FIGURE 3.
Neo N-terminal sequences after proteolytic cleavage as determined by both Edman degradation and mass spectrometry. A, schematic representation of the human MUC2-C corresponding to the region directly after the PTS domain (amino acids 4198–5179) flanked at the N-terminal side by GFP and the Myc tag. The two cleavage sites identified by Edman sequencing at ↓TTGLR and ↓QAVAL are highlighted, plus the peptide TTGLR and QAVALPYK as confirmed by mass spectrometry analysis. The localization of the epitope for the anti-MUC2C2 polyclonal antibody is highlighted at the far C terminus. B, fragmentation spectra of the TMPP-labeled peptide TTGLR. The MUC2 C terminus was incubated with P. gingivalis secretions and labeled using TMPP specifically derivatizing the newly formed N terminus. Mass spectrometry analysis identified the peptide TTGLR labeled at its N terminus, indicating proteolytic cleavage at this site. The TMPP-CHCO+ reporter ion is indicated in red, b-ions are in blue, and y-ions are in red.