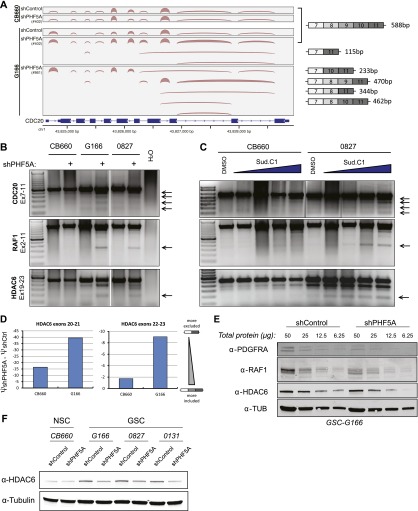
Figure 3.
Loss of PHF5A results in splicing defects in GSCs but not NSCs. (A) Select genes important for cell cycle progression, such as CDC20, display broad splicing defects following PHF5A knockdown. Plot illustrates the density of RNA-seq reads crossing splice junctions and was created with IGV (Robinson et al. 2011). Aberrant isoforms lacking constitutive exons appear following knockdown of PHF5A with two distinct shRNAs. cDNA base pair sizes indicate the expected product size of each isoform using the cdc20 primers indicated in C and D. (B) RT–PCR of RNA isoforms of example genes after PHF5A knockdown. PCR products were generated using primers in the indicated exons of each gene. Arrows indicate splicing products specifically induced in GSCs after PHF5A knockdown. (C) RT–PCR as in B using RNA from cells treated for 24 h with 0.5, 1, 2, or 4 μM SudC1. (D) qRT–PCR using primers designed to specifically recognize the splice junctions between consecutive and nonconsecutive exons was performed to determine ψ-values for inclusion of potentially skipped exons in HDAC6 identified above. The change in inclusion rate after PHF5A knockdown is presented. (E) Serial dilutions of GSC lysate with or without PHF5A knockdown were run as Western blots and probed with antibodies specific to example genes with predicted missplicing events. (F) Western blot of GSCs and NSCs with or without PHF5A knockdown probed for levels of the frequently misspliced protein HDAC6. See also Supplemental Figure S3.