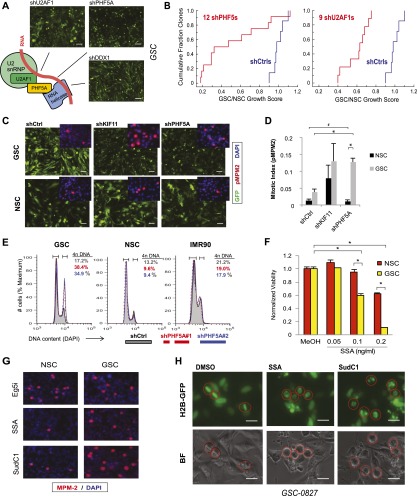
Figure 4.
PHF5A and its binding partners are differentially required for GSC expansion, and their inhibition triggers G2/M cell cycle arrest in GSCs but not NSCs. (A) Model of PHF5A splicing interactions based on previous research. Knockdown of known PHF5A-binding partners in the spliceosome recapitulates the PHF5A knockdown phenotype, shown. Bar, 50 μm. (B) Cumulative probability plot of multiple shRNAs scoring as cell-lethal versus their GSC to NSC viability ratio. (C) Fluorescence microscopy images of GSCs or NSCs expressing constitutive GFP and PHF5A knockdown or control constructs. (Inset) Immunofluorescent images of phospho-MPM2, indicative of mitotic activity, in GSCs and NSCs depleted for PHF5A. Bar, 100 μm. (D) Quantification of phospho-MPM2 staining in GSCs and NSCs infected with control or shPHF5A virus. (*) P-value < 0.001; (#) insignificant P-value = 0.5. (E) Cellular DNA content as measured by DAPI staining and FACS analysis in GSC, NSC, and IMR90 normal fibroblast cultures with or without PHF5A knockdown. The percentage of cells in each sample with 4n DNA content is shown, indicative of cells that have completed DNA replication in S phase. (F) Viability of GSCs or NSCs treated with increasing doses of SSA. (*) P-value < 0.0003. (G) Immunofluorescent images of phospho-MPM2 in GSCs and NSCs treated with SSA or SudC1. (H) Fluorescent and light images of GSCs expressing Histone 2B-GFP fusion protein, which marks DNA. Cells were pretreated with SSA or SudC1 for 22 h before addition of MG132 for an additional 2 h to arrest dividing cells in metaphase. Circles mark the same cell in corresponding images. See also Supplemental Figure 4 and Supplemental Movies S1–S3.