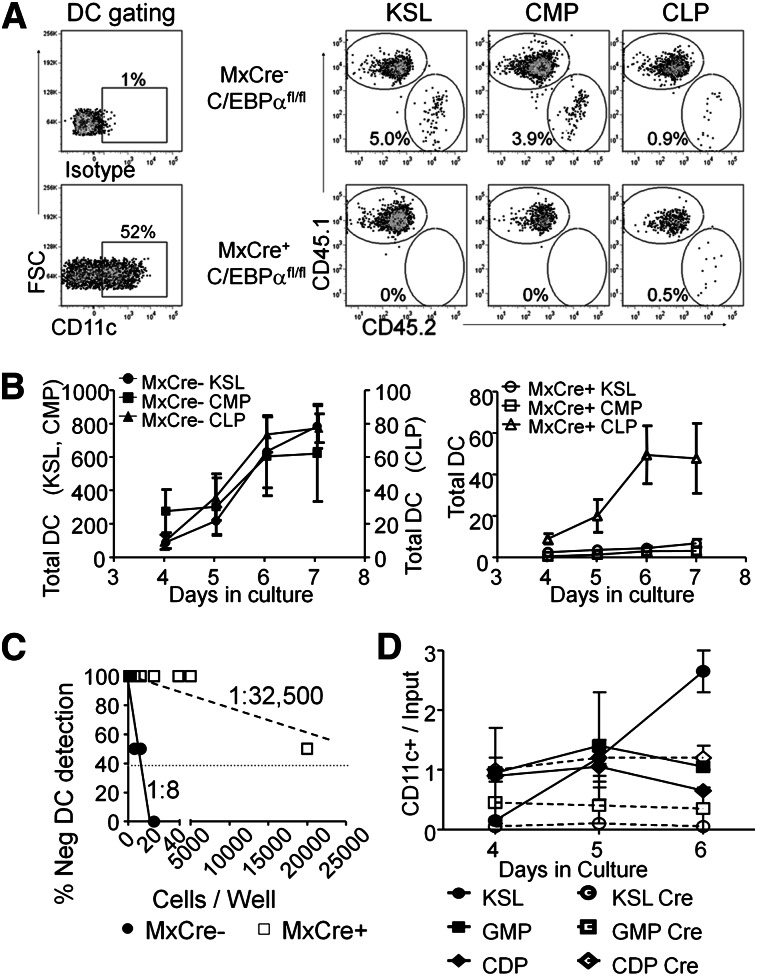
Figure 3.
Stage-specific need for C/EBPα in DC progenitors. (A) KSL, CMPs, and CLPs were isolated from the BM of MxCre+ C/EBPαfl/fl or C/EBPαfl/fl mice 2 weeks after 3 poly(IC) injections. 500 progenitor cells were cocultured with 104 Ly5.1+ WBM cells in the presence of Flt3L (200 ng/mL). The resulting cultures were analyzed by flow cytometry for Ly5.1, Ly5.2, MHCII, and CD11c after days 3 to 7. In addition, CD45.1+ cells were subjected to the same culture conditions that were used as controls for efficiency of DC differentiation. Day 5 FACS plots are shown as representative data from triplicate wells of 3 independent experiments. (B) The number of DCs generated in each culture was calculated using cell counts and flow cytometry over days and presented as the number of total DCs (average of 3 wells). On the left are sorted populations from C/EBPα fl/fl, with the DC numbers for KSL and CMPs shown on the left axis and CLP production on the right axis. The graphs on the right are from the same progenitors but sorted from MxCre+ C/EBPαfl/fl mice. Data are presented as mean ± SD of one such experiment that was repeated 3 times. (C) Cloning efficiency was assessed by limiting dilution series of sorted CD45.2+ MxCre+ C/EBPαfl/fl or C/EBPαfl/fl Lin– cKit+ progenitors cocultured with Ly5.1+ Lin– cKit+ BM cells in the presence of Flt3L(200 ng/mL). Cultures were analyzed on day 8 for the presence of CD45.2+ CD11c+ cells. Data are representative of 2 such experiments. (D) The progenitor populations KSL, GMP, and CDP were isolated from the BM of C/EBPαfl/fl mice. These progenitor cells were infected with retrovirus carrying an empty vector with GFP reporter or Cre with GFP reporter. These cells were then cultured with sorted CD45.1+ Lin– cKit+ BM cells in the presence of Flt3L. Cultures were assessed for CD45.2+ CD11c+ GFP+ cells at days 4, 5, and 6. The plot shows a yield of CD11c+ cells per input GFP+ progenitor over time. Data are representative of 2 such experiments.