Fig. 2. R-2HG can serve as an EglN cosubstrate.
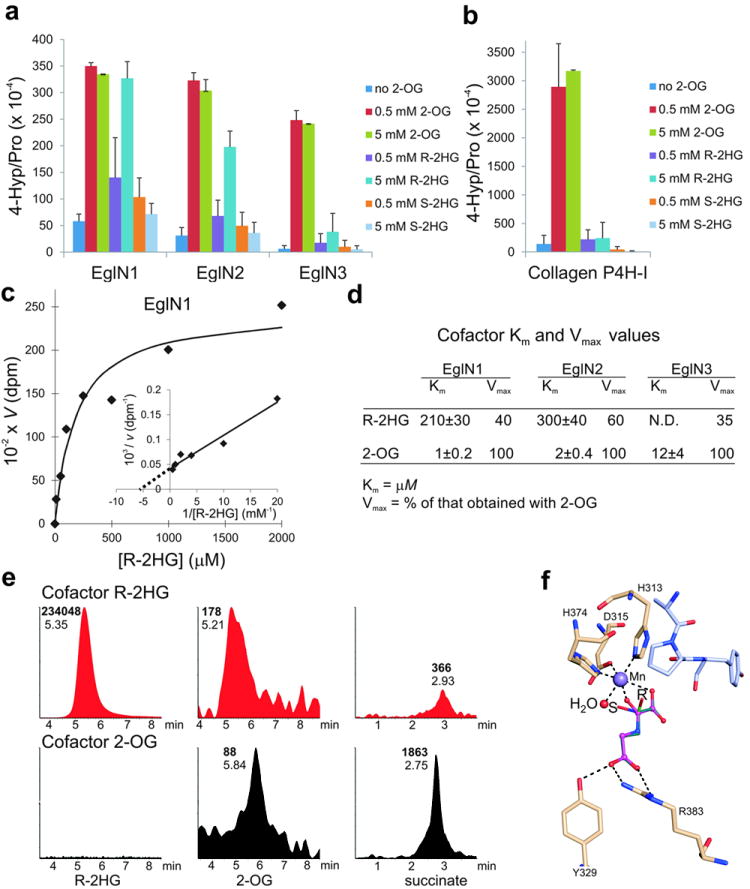
a, b, In vitro prolyl 4-hydroxylation assays conducted with recombinant EglNs (a) and collagen P4H-I (b) in the presence of the indicated amounts of 2-OG or 2HG. L-[2,3,4,5-3H]proline-labeled HIF1α oxygen-dependent degradation domain (ODDD) (a) and [14C]proline-labeled protocollagen (b) were used as substrates. Enzymes were produced in insect cells using baculoviruses and affinity-purified. Error bars, s.d.; n = 3-4.
c, d, Km and Vmax values for R-2HG for EglN family members. Km and Vmax values for 2-OG 21 are included for comparison.
e, LC-MS analysis of succinate, 2-OG and R-2HG from enzymatic reactions with EglN1, HIF1α oxygen-dependent degradation domain (ODDD) polypeptide and either 5 mM R-2HG (red) or 80 μM 2-OG (black) as cofactors. Numbers next to each peak indicate elution times (plain font) and peak areas (bold font). No peaks above background were detected in samples in which 2-OG and R-2HG were both omitted (data not shown).
f, Model of R-2HG (green) and S-2HG (cyan) bound to the active site of EglN1. N-oxalylglycine (magneta) bound in the original structure 29 is shown for comparison. The active site water molecule, which has been shown to be the O2 binding site 30, is shown in red and the peptide substrate in light blue. Hydrogen bonds are indicated by dash lines.
