Fig. 4.
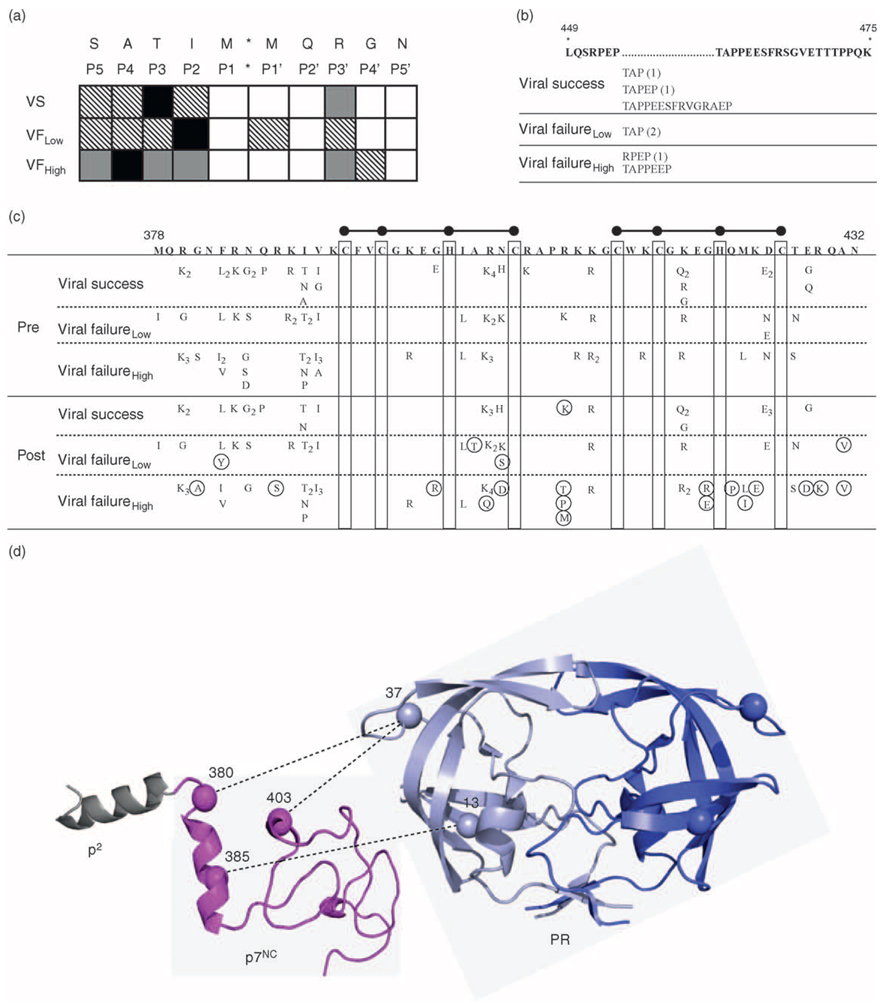
(a) Diversity in p2/p7NC cleavage site. Patients are stratified by therapy response: viral success, viral failure with low fit viruses, or viral failure with high fit viruses. Amino acid sequence of HIVHXB2 cleavage site is shown on top line with cleavage site position number shown below each respective amino acid. Black box, polymorphism present in more than 50% of patients; gray box, polymorphism present in 26–50% of patients; hatched box, polymorphism present in 1–25% of patients; white box, no polymorphism. (b) Length variation in p6. Amino acid sequence and position numbers of p6 (HIVHXB2) are shown on top, with regions containing length variation marked with (…). The amino acid compositions of insertions within each response group (number of patients) are shown below each region of length variation. (c) Variability in p7NC. Amino acid sequence and position numbers of p7NC (HIVHXB2) are shown on top. Ball and sticks represent the zinc finger motifs. Polymorphisms present prior to therapy are shown at the top, and polymorphisms after 24 weeks of therapy are shown on the bottom, with subscripts representing the number of patients with a particular polymorphism. Mutations that accumulate in p7NC following 24 weeks of therapy are circled. (d) Model of p2, p7NC, and protease, with specific covarying positions in protease and p7NC noted. Amino acid residues in p7NC are numbered according to HIVHXB2, and amino acid residues in protease are numbered according to position within protease. Dashed lines indicate pretherapy covariation between positions in protease and positions in p7NC.