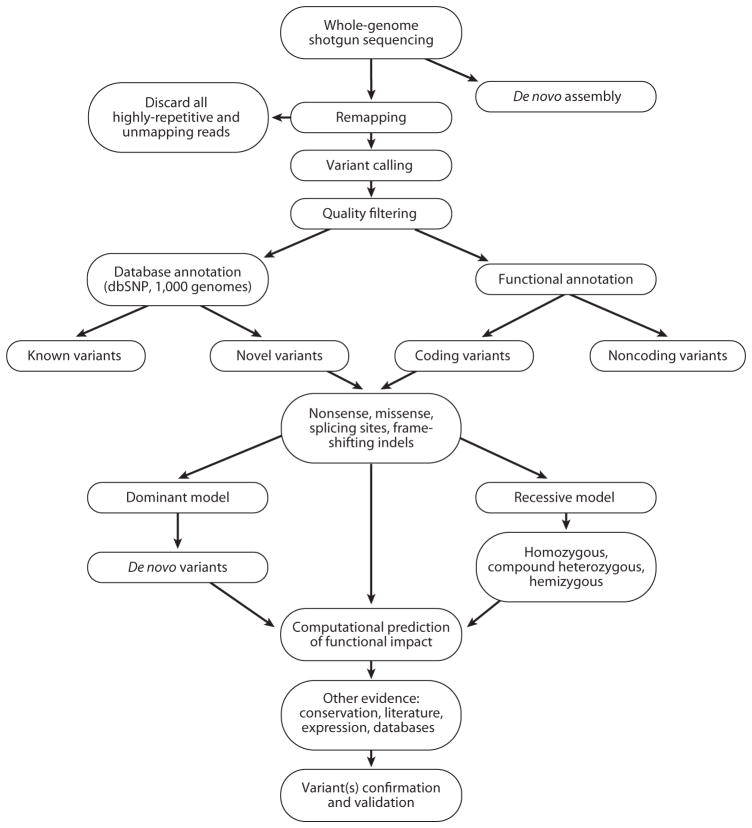
Figure 4.
Schematic workflow of whole-genome/exome sequencing data analysis. After sequencing, the sequence reads are mapped and aligned against the human reference genome assembly in order to obtain a list of variants at every position that does not match the reference. Quality filters are applied to obtain high-quality variant calls. Various filtering criteria are applied to prioritize the candidate variants. Most variants will be excluded because they are known, meaning that they are already in variation databases, such as the database of single nucleotide polymorphisms (dbSNP), The 1000 Genomes Project database, etc. The focus is mainly on novel variants, which can be tiered in functional classes according to their annotation. For coding variants, priority is given to nonsense, frameshifting, splice-site, and then missense mutations. Computational prediction of the functional impact of these variants can also help prioritize candidate mutations. Based on the characteristics of the trait or disease of interest, variants can be examined under a dominant or recessive model. Additional confirmation through other resources can strengthen the hypotheses of the functional significance of identified variants. Genetic and functional confirmation of the candidate disease-causing variants is the final, most important step.