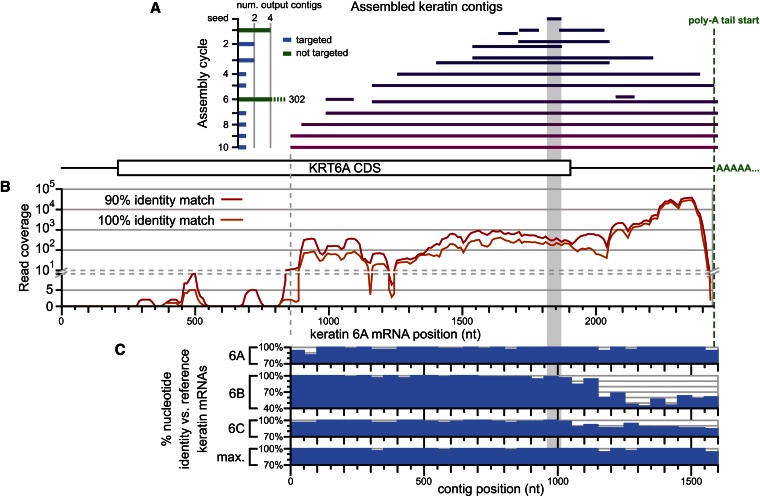
Figure 3.
De novo assembly of the keratin 6A mRNA from a transcriptome dataset. (A) Contigs from each step of a 10-cycle PRICE assembly aligned to the keratin 6A reference sequence (NM_005554.3) by BLASTn (Altschul et al. 1990). The seed sequence is a single 54nt read from the paired-end transcriptome dataset (Arron et al. 2011) (dark blue; see Materials and Methods); the later contigs (purple) include poly-A tail sequence not included in the reference sequence. Left: the total number of output contigs generated in each cycle, shown as a histogram of blue or green bars for cycles that were or were not explicitly targeted (using the –target flag; see Materials and Methods) to the seed sequence, respectively. (B) Read coverage from the 54nt paired-end read dataset determined by mapping to the keratin 6A reference. Units are the number of reads overlapping each nucleotide, averaged across nonoverlapping 10nt windows. Coverage is shown requiring 90% (red) or 100% (orange) nucleotide identity between the read and the reference. (C) Identity of the PRICE contig vs. the reference keratin 6A sequence, as well as the human keratin 6B and 6C isoforms (NM_005555.3 and NM_173086.4, respectively) for nonoverlapping 50nt windows and including the poly-A tail sequence. Bottom, the maximum % identity for each 50nt contig window to the three keratin 6 isoforms.