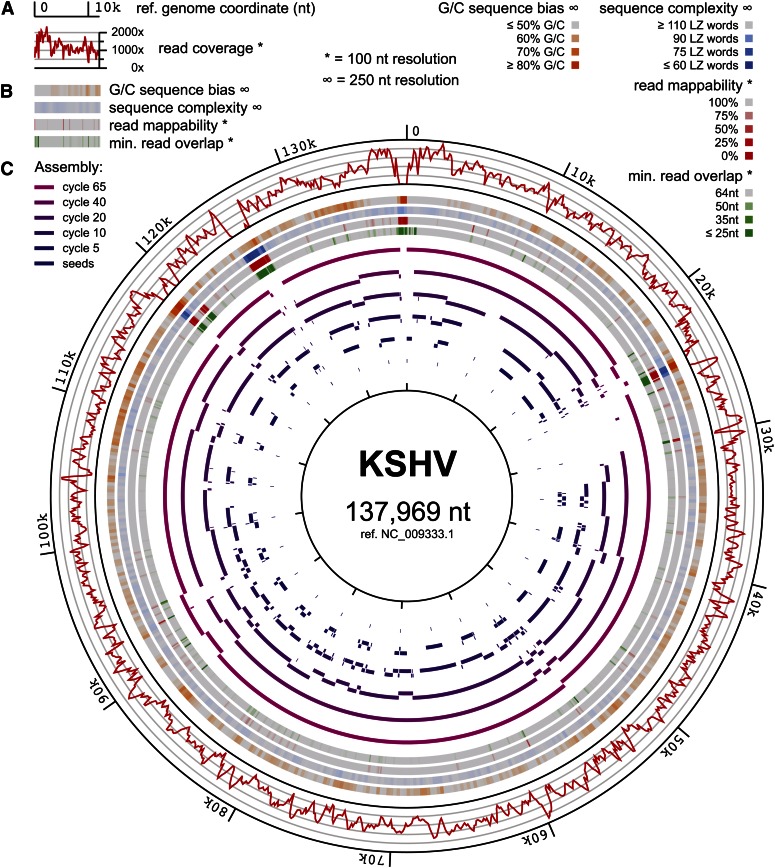
Figure 4.
De novo assembly of the BCBL-1 strain of the KSHV. (A) Read coverage from the 65nt paired-end read dataset determined by mapping to the KSHV reference genome (Rezaee et al. 2006) (NC_009333.1) with BLASTn (Altschul et al. 1990), requiring 90% identity. Units are the number of reads overlapping each nucleotide, averaged across nonoverlapping 100nt windows. (B) Heat maps indicating the percent of nucleotides that are G/C in nonoverlapping 250nt windows (orange), LZW sequence complexity (Welch 1984) of nonoverlapping 250nt sequences (blue; see Materials and Methods), read mappability as determined by mapping every overlapping 65mer from the genome using the same method as in (A) and averaging the coverage over a 100nt window (red), and the minimum overlap between adjacently mapping reads across each 100nt window, measured as the minimum value across all reads with 3′ ends in the window, measuring the maximum overlap with all reads mapped 3′ of the given read (green). (C) Contigs from selected steps of a 65-cycle PRICE assembly aligned to the reference genome. Seed sequences of 65nt are shown as the innermost ring (dark blue), followed by intermediate contigs aligned to the reference genome by BLASTn (Altschul et al. 1990), with the final contigs aligned to the reference genome by the Smith-Waterman method (Smith and Waterman 1981) shown on the outer ring (purple).