Figure 2.
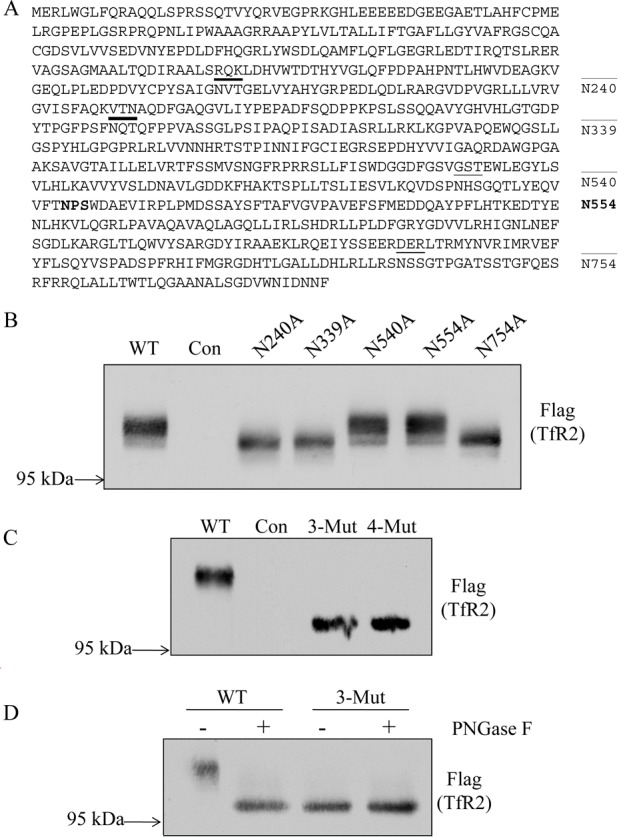
Identification of N-linked glycosylation sites in hTfR2. (A) Schematic representation of the N-linked glycosylation sites on hTfR2 protein sequence. Asn 240, 339, 540, and 754 are predicted to be glycosylated (underlined). Asn 554 (bold) has an NPS/T motif and was used as a negative control because this Asn could not be glycosylated. (B) Western blot analysis of cell lysates from HEK 293 cells transiently transfected with empty vector (pcDNA3, Con) or hTfR2-FLAG expression vectors encoding either the wild type (WT) or N-glycosylation site mutants (N240A, N339A, N540A, and N754A). Positions 240, 339, and 754 are identified as being glycosylated, but position 540 is not. N554A was used as a negative control. (C) Western blot analysis of cell lysates from HEK 293 cells transiently transfected with empty vector (pcDNA3, Con), WT hTfR2 (WT), the hTfR2 nonglycosylated triple mutant (N240/339/754A, 3-Mut), or the quadruple mutant (N240/339/540/754A, 4-Mut). (D) HEK 293 cells transfected with empty vector (pcDNA3, Con), WT hTfR2 (WT), or the hTfR2 nonglycosylated mutant (N240/339/754A, 3-Mut) were harvested, and cell lysates were incubated with or without PNGase F before Western blotting. The samples were electrophoresed on a 12 cm 10% polyacrylamide gel for 24 h to ensure greater separation, transferred to nitrocellulose, and probed with anti-Flag antibody for TfR2. The data represent three independent experiments.
