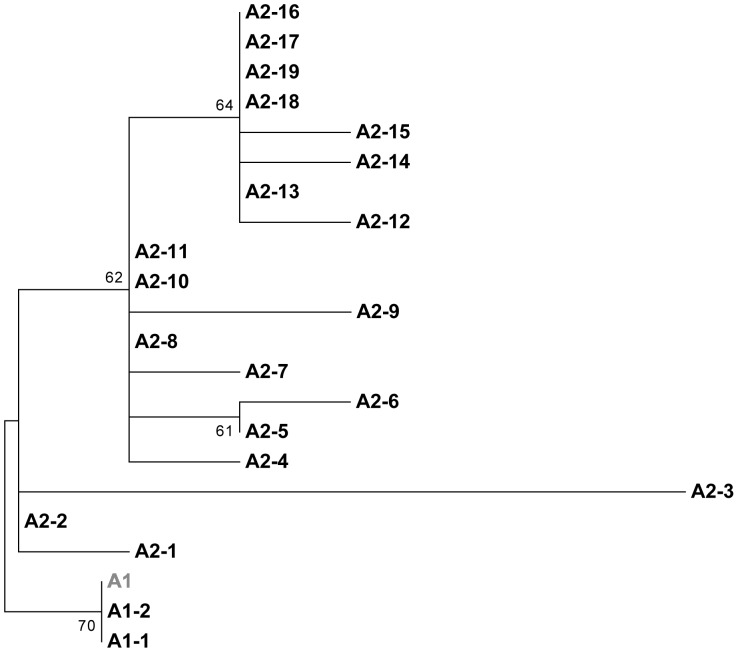
Figure 2. HPV11 LCR phylogenetic tree from Australia and other geographical regions.
A maximum likelihood (ML) tree was inferred from molecular phylogenetic analysis of 94 aligned LCR sequences using MEGA5 based on the Tamura-Nei model with 500 bootstrap replicates [66]. These 94 sequences were derived from Australia (KC329850–KC329871), Slovenia (FN870625–FN870687), China (EU918768), Hungary (HE574701, HE574705, FR872717) and Thailand (JQ773408–JQ773412). The reference sequence (M14119) from sublineage A1 is shown in grey. The 21 variants are named by the lineage they cluster to. Sublineage A1 was comprised of one Slovenian variant (A1–1) and one geographically shared variant (A1–2). Sublineage A2 was comprised of 6 Australian (A2–3, A2–6, A2–13, A2–14, A2–16, A2–17), Slovenian(A2–1, A2–7, A2–10, A2–12, A2–15), Hungarian (A2–2), Thai(A2–9), and geographically shared (A2–4, A2–5, A2–8, A2–11, A2–18, A2–19) variants. In cases where sequences were identical only one sequence from that country was use to represent each variant. In seven instances variants from different geographical regions were also identical, again one sequence was used to represent each variant; A1–2 (China and Slovenia), A2–4 (Hungary and Thailand), A2–8 (Australia, Hungary and Slovenia), A2–5, A2–11, A2–18 (Australia and Slovenia), A2–19 (Australia, Slovenia and Thailand).