Abstract
Specialization makes resource use more efficient and should therefore be a common process in animal evolution. However, this process is not as universal in nature as one might expect. Our study shows that Sphecodes (Halictidae) cuckoo bees frequently change their host over the course of their evolution. To test the evolutionary scenario of host specialization in cuckoo bees, we constructed well-supported phylogenetic trees based on partial sequences of five genes for subtribe Sphecodina (Halictini). We detected up to 17 host switches during Sphecodes evolution based on 37 ingroup species subject to mapping analysis of the hosts associated with the cuckoo bee species. We also examine the direction of evolution of host specialization in Sphecodes using the likelihood ratio test and obtain results to support the bidirectional evolutionary scenario in which specialists can arise from generalists, and vice versa. We explain the existence of generalist species in Sphecodes based on their specialization at the individual level, which is recently known in two species. Our findings suggest flexible host choice and frequent host switches in the evolution of Sphecodes cuckoo bees. This scenario leads us to propose an individual choice constancy hypothesis based on the individual specialization strategy in cuckoo bees. Choice constancy has a close relationship to flower constancy in bees and might be an extension of the latter. Our analysis also shows relationships among the genera Microsphecodes, Eupetersia, Sphecodes and Austrosphecodes, a formerly proposed Sphecodes subgenus. Austrosphecodes species form a basal lineage of the subtribe, and Microsphecodes makes it paraphyletic.
Introduction
If there were a species that could utilize all available resources equally well, it would certainly be the most successful species on Earth. In fact, there is no such species because adaptation to an extremely wide spectrum of different resources is impossible. According to Van Valen's Red Queen hypothesis [1], resource specialization seems to be a necessary condition to keep pace in an evolutionary race between interacting species (an “arms race” [2]). Apparently, there are two opposing pressures: it would be beneficial to utilize a broad resource spectrum, but specialization is necessary for effective resource utilization. Therefore, we can find resource specialists as well as resource generalists, and each strategy has some advantages and disadvantages.
Specialists could be favored over generalists because they do not face the problems of antagonistic adaptations to different resources; thus, specialists can respond to counter-adaptations more effectively [3]. They can also process information more easily during the search for suitable resources [4] and occupy a less competitive environment [5]. Finally, a new advantageous allele that allows more efficient resource utilization can spread faster in specialized species [6], [7].
Generalists, by contrast, could benefit from greater resource availability and therefore require less investment in resource acquisition [5], [8]. It is unclear whether the generalist strategy is simply a temporary condition in evolutionary terms that will inevitably lead to specialization as the possible stable strategy.
Alternatively, perhaps specialists are prone to extinction because their narrow adaptation inhibits the utilization of other resources [3]. According to this hypothesis, specialization is an evolutionary dead end [9]. Cospeciation, or “missing the boat” (i.e., species utilize only one of two new resources established through speciation) [10] are the routes through which specialists can switch to new resources. Switching to a new unrelated resource is impossible under these assumptions.
However, the dead-end hypothesis is not consistent with the results of many empirical studies, as summarized in Hoberg's and Brooks' review [11] of parasites. Host switches with no or weak cospeciation with hosts are also commonly documented in herbivorous insects [12], [13], [14], [15], [16], [17], [18]; thus, generalists can evolve from specialist species [14], [15]. A decreasing trend of host specificity has also been documented in parasites, such as fleas [19] or Monogenea [20].
The ecological fitting hypothesis explains the results of most empirical studies. According to this hypothesis, a common evolutionary history is not necessary for the mutual adaptation of two species. The reason why two species are adapted to each other results from the coincidence of their compatibility regarding important characters [21]. The switch to a new resource is conditioned by potential fitness, which the species would have in a new situation [22]. Another explanation reflects the fact that some resources do not require special adaptations for their utilization. Such conditions allow easy switches to new resources [23], especially switches between ecologically similar resources [11].
Resource specialization appears to be a very complex process, which is potentially shaped by many factors. An example is provided by pollen specialization, until now, the only well studied case of resource specialization in bees. There are both pollen generalists and pollen specialists [24]. Theoretically, the generalist strategy would be the more advantageous because it would allow all types of pollen resources to be utilized. Nevertheless, this strategy is not possible because the choice of host flower is physiologically or neurologically constrained. The pollen of certain plants may be toxic to some bee species, resulting in a physiological constraint [25], [26]. Even the larvae of generalist species are unable to develop on the pollen collected by another generalist species [27]. A neurological constraint arises because bees have limited memory and learning capacities with respect to shape and color [28], [4]. Therefore, the host flowers within bee species are usually similar [29]. A pollen specialist would, in theory, be better adapted to these conditions; however, pollen generalist species of bees exist, and the generalist strategy seems to be the derived condition [30], [31]. Generalist species may be successful because their pollen-collecting behavior resembles that of the specialist. Generalist species focus on a single host plant species before switching to new host flowers after a couple of foraging visits [32]. This phenomenon, called flower constancy, may allow bees to overcome neurological constraints [32]. Similarly, the specialized strategy could be more flexible than it may first appear: in several pollen specialized bee species, host switching has been observed in the absence of their usual flower host [33], [34]. Such bee species may have one essential host but be able to utilize other hosts.
Here, we address the cuckoo bees, a parasitic group of bees that do not collect pollen. Cuckoo bees neither build their own nest nor provision brood cells. Instead, these bees search for nests of other bee species and lay eggs in their cells. The host provisions of pollen are used as nutrition by the offspring of the cuckoo bee. Cuckoo behavior is relatively common in bees. Obligatory cuckoo species are found in four of the seven bee families [24], and 27 independent origins of this behavior are now identified in bees [24], [35], [36]. Most cuckoo bee species are specialists, having one or a few closely related hosts. Nevertheless, generalist species with many hosts of different genera and families are also known [37], [38]. We investigated the evolution of host specificity in the genus Sphecodes Latreille in this study because the host species are relatively well studied and both specialist and generalist species occur within this genus [39]. Moreover, two generalist Sphecodes species (S. monilicornis and S. ephippius) are specialized at the individual level: individual females repeatedly visit the nests of the same host species, but different females visit different hosts [38]. Sphecodes belongs to the tribe Halictini, subtribe Sphecodina, a group consisting of 249 species worldwide. These species live in all ecosystems and are most likely all parasitic [24]. Microsphecodes Eickwort and Stage, Eupetersia Blüthgen and Ptilocleptis Michener are the other genera of this subtribe [24]. Michener [40], [24] suspects that Eupetersia and Ptilocleptis are basal taxa of Sphecodina and recognizes two subgenera in Sphecodes: Sphecodes sensu stricto and Austrosphecodes Michener. Nevertheless, no phylogenetic study has been conducted to date, making the evolutionary relationships within this subtribe unclear.
Using the cuckoo bees as a model, this paper seeks to enhance our understanding of the general principles of resource specialization. Therefore, our goals were (1) to develop a robust phylogeny of the subtribe Sphecodina and discuss the result, (2) to test whether cuckoo bees of the genus Sphecodes have evolved in concert with their host species and whether host switches are rare or common, (3) to test whether generalism is an ancestral strategy in cuckoo bees and whether reversals in host specialization have occurred, (4) to determine whether specialization at the individual level is just a unique and relatively unstable strategy in cuckoo bee evolutionary history and whether this generalist strategy can lead to species diversification through specialization on new hosts and (5) to develop a hypothesis of resource specialization based on our results.
Methods
Materials and data sets
The complete data set comprises 107 specimens of 48 species from the subtribe Sphecodina and 20 species from the subfamilies Rophitinae, Nomiinae, Nomioidinae and Halictinae (tribes Augochlorini, Caenohalictini and Halictini) used as outgroups. Most of the sequences are newly developed for this study. The sequences of the outgroup taxa and some Sphecodes were acquired from the NCBI database (Table 1). A reduced data set including only one specimen per species and all outgroup species was used for our primary analysis and for the reconstruction of ancestral character states. The complete list of specimens, locality data and GenBank accession numbers are listed in Table 1. Voucher specimens of taxa used for the first time were deposited in the J. Straka collection (Charles University in Prague). Most bee specimens were preserved in 96% EtOH; several pinned specimens were also used. The nomenclature follows Michener [24] and Bogusch and Straka [39].
Table 1. Complete list of specimens, locality data and Genbank accession numbers of sequences.
| GenBank Accession numbers | |||||||
| Taxon, No. | Collection country | Align1 | 28S | COI | EF1 | LWR | WG |
| Agapostemon tyleri | – | x | AY654506.1a | AF102835.1b | AF140320.1c | AY227940.1d | AY222577.1e |
| Augochloropsis metallica | – | x | GU320093.1f | – | AF140315.1c | AY227934.1d | AY222571.1e |
| Austrosphecodes sp. 1 | Peru | x | JX256745 | JX256648 | JX256448 | JX256782 | JX256546 |
| Austrosphecodes sp. 12 | Argentina | x | JX256746 | JX256649 | JX256449 | JX256783 | JX256547 |
| Austrosphecodes sp. 13 | Argentina | x | JX256747 | JX256650 | JX256450 | JX256784 | JX256548 |
| Austrosphecodes sp. 14 | Argentina | x | JX256748 | JX256651 | JX256451 | JX256785 | JX256549 |
| Conanthalictus wilmattae | – | x | AY654508.1a | – | AF435378.1g | AY227916.1d | AY222553.1e |
| Dieunomia nevadensis | – | x | DQ060852.1h | – | AF435396.1g | AY227931.1d | AY222568.1e |
| Dufourea novaeangliae | – | x | – | FJ582211.1i | AF435384.1g | AY227919.1a | AY222556.1e |
| Eupetersia seyrigi | – | x | – | – | EU203259.1e | EU203287.1e | EU203228.1e |
| Eupetersia sp. 1 | Angola | x | JX256749 | JX256652 | JX256452 | JX256786 | JX256550 |
| Halictus ligatus | – | x | – | AF102840.1b | AF140299c | AY455895.1j | AY455899.1j |
| Halictus quadricinctus | – | x | – | AF438422.1k | AF140334.1c | AY227956.1d | AY222592.1e |
| Lasioglossum calceatum | – | x | – | AF103980.1b | AF435385.1g | AF448877.1l | AY222608.1e |
| Lasioglossum florale | – | x | – | AF103955.1b | AF264792.1m | AY227966.1d | AY222602.1d |
| Lasioglossum hybodinum | – | x | GU320096.1f | AF104660.1b | AF264857.1m | AY227963.1d | AY222599.1d |
| Lasioglossum lustrans | – | x | – | AF104643.1b | AF435388.1g | AF448904.1l | AY222609.1e |
| Lasioglossum zephyrum | – | x | – | AF103974.1b | AF435379.1g | AF448918.1l | AY222607.1e |
| Lasioglossum zonulum | – | x | – | AF104658.1b | AF264855.1m | AY227969.1d | AY222606.1d |
| Microsphecodes sp. 10 | Venezuela | x | JX256750 | JX256653 | JX256453 | JX256787 | JX256551 |
| Microsphecodes sp. 15 | Bolivia | x | JX256751 | JX256654 | JX256454 | JX256788 | JX256552 |
| Nomioides facilis | – | x | AY654511.1a | – | AF435394.1g | AY227929.1d | AY222566.1d |
| Patellapis abessinica | – | x | GU320097.1f | – | EU203267.1e | EU203295.1e | EU203236.1e |
| Penapis penai | – | x | AY654513.1a | – | AF435401.1g | AY227921.1d | AY222558.1e |
| Ruizantheda mutabilis | – | x | GU320094.1f | – | AF435406.1g | AY227949.1d | AY222586.1e |
| Sphecodes albilabris 1 | Czech Republic | x | JX256752 | JX256655 | JX256455 | JX256789 | JX256553 |
| Sphecodes albilabris 2 | Czech Republic | – | – | JX256656 | JX256456 | – | JX256554 |
| Sphecodes albilabris ssp. 3 | Spain | – | – | JX256657 | JX256457 | – | JX256555 |
| Sphecodes alternatus 1 | Hungary | x | JX256753 | JX256658 | JX256458 | JX256790 | JX256556 |
| Sphecodes alternatus 2 | Italy | – | – | JX256659 | JX256459 | – | JX256557 |
| Sphecodes autumnalis | – | x | – | FJ582459.1i | EU203256.1e | – | EU203225.1e |
| Sphecodes cf. dusmeti | Iran | x | JX256758 | – | JX256468 | JX256793 | JX256566 |
| Sphecodes clematidis | – | x | – | FJ582469.1i | EU203257.1e | – | EU203226.1e |
| Sphecodes confertus | – | x | – | FJ582472.1i | EU203258.1e | – | EU203227.1e |
| Sphecodes crassus 1 | Czech Republic | x | JX256756 | JX256663 | JX256463 | – | JX256561 |
| Sphecodes crassus 2 | Slovakia | – | – | JX256664 | JX256464 | – | JX256562 |
| Sphecodes crassus 4 | Czech Republic | – | – | JX256665 | JX256465 | – | JX256563 |
| Sphecodes crassus 6 | Switzerland | – | – | JX256666 | JX256466 | – | JX256564 |
| Sphecodes cristatus | Slovakia | x | JX256757 | JX256667 | JX256467 | JX256792 | JX256565 |
| Sphecodes croaticus | Czech Republic | x | JX256755 | JX256662 | JX256462 | – | JX256560 |
| Sphecodes ephippius 1 | Hungary | x | JX256759 | JX256668 | JX256469 | JX256794 | JX256567 |
| Sphecodes ephippius 2 | Czech Republic | – | – | JX256673 | JX256474 | – | JX256572 |
| Sphecodes ephippius 3 | Czech Republic | – | – | JX256674 | JX256475 | – | JX256573 |
| Sphecodes ephippius 4 | Czech Republic | – | – | JX256675 | JX256476 | – | JX256574 |
| Sphecodes ephippius 8 | Czech Republic | – | – | JX256676 | JX256477 | – | JX256575 |
| Sphecodes ephippius 9 | Hungary | – | – | JX256677 | JX256478 | – | JX256576 |
| Sphecodes ephippius 14 | Italy | – | – | JX256669 | JX256470 | – | JX256568 |
| Sphecodes ephippius 15 | Iran | – | – | JX256670 | JX256471 | – | JX256569 |
| Sphecodes ephippius 16 | Bulgaria | – | – | JX256671 | JX256472 | – | JX256570 |
| Sphecodes ephippius 17 | Czech Republic | – | – | JX256672 | JX256473 | – | JX256571 |
| Sphecodes ferruginatus 1 | Czech Republic | x | JX256760 | JX256678 | JX256479 | JX256795 | JX256577 |
| Sphecodes ferruginatus 2 | Czech Republic | – | – | JX256679 | JX256480 | – | JX256578 |
| Sphecodes geoffrellus 1 | Czech Republic | x | JX256761 | JX256680 | JX256481 | JX256796 | JX256579 |
| Sphecodes geoffrellus 2 | Czech Republic | – | – | JX256681 | JX256482 | – | JX256580 |
| Sphecodes geoffrellus 7 | Czech Republic | – | – | JX256682 | JX256483 | – | JX256581 |
| Sphecodes gibbus 1 | Czech Republic | x | JX256762 | JX256683 | JX256484 | JX256797 | JX256582 |
| Sphecodes gibbus 2 | Czech Republic | – | – | JX256685 | JX256486 | – | JX256584 |
| Sphecodes gibbus 3 | Czech Republic | – | – | JX256686 | JX256487 | – | JX256585 |
| Sphecodes gibbus 4 | Slovakia | – | – | JX256687 | JX256488 | – | JX256586 |
| Sphecodes gibbus 5 | Czech Republic | – | – | JX256688 | JX256489 | – | JX256587 |
| Sphecodes gibbus 6 | Czech Republic | – | – | JX256689 | JX256490 | – | JX256588 |
| Sphecodes gibbus 7 | Czech Republic | – | – | JX256690 | JX256491 | – | JX256589 |
| Sphecodes gibbus 10 | Iran | – | – | JX256684 | JX256485 | – | JX256583 |
| Sphecodes hyalinatus 3 | Czech Republic | – | – | JX256691 | JX256492 | – | JX256590 |
| Sphecodes hyalinatus 4 | Bulgaria | x | JX256763 | JX256692 | JX256493 | JX256798 | JX256591 |
| Sphecodes longuloides | Spain | x | JX256764 | JX256693 | JX256494 | JX256799 | JX256592 |
| Sphecodes longulus 1 | Slovakia | x | JX256765 | JX256694 | JX256495 | JX256800 | JX256593 |
| Sphecodes longulus 2 | Czech Republic | – | – | JX256695 | JX256496 | – | JX256594 |
| Sphecodes longulus 3 | Italy | – | – | JX256696 | JX256497 | – | JX256595 |
| Sphecodes majalis | Czech Republic | x | JX256767 | JX256703 | JX256505 | JX256803 | JX256603 |
| Sphecodes marginatus 6 | Tunisia | – | – | JX256698 | JX256499 | – | JX256597 |
| Sphecodes marginatus 7 | Tunisia | – | – | JX256699 | JX256500 | – | JX256598 |
| Sphecodes marginatus 10 | Tunisia | x | JX256766 | JX256697 | JX256498 | JX256801 | JX256596 |
| Sphecodes miniatus 2 | Slovakia | – | – | JX256700 | JX256501 | – | JX256599 |
| Sphecodes miniatus 4 | Czech Republic | – | – | JX256701 | JX256502 | – | JX256600 |
| Sphecodes miniatus 7 | Switzerland | – | – | JX256702 | JX256503 | – | JX256601 |
| Sphecodes miniatus 9 | Slovakia | x | – | – | JX256504 | JX256802 | JX256602 |
| Sphecodes monilicornis 1 | Czech Republic | x | JX256768 | JX256704 | JX256506 | JX256804 | JX256604 |
| Sphecodes monilicornis 2 | Czech Republic | – | – | JX256706 | JX256508 | – | JX256606 |
| Sphecodes monilicornis 4 | Czech Republic | – | – | JX256707 | JX256509 | – | JX256607 |
| Sphecodes monilicornis 7 | Czech Republic | – | – | JX256708 | JX256510 | – | JX256608 |
| Sphecodes monilicornis 8 | Italy | – | – | JX256709 | JX256511 | – | JX256609 |
| Sphecodes monilicornis 9 ssp. cephalotes | Turkey | – | – | JX256710 | JX256512 | – | JX256610 |
| Sphecodes monilicornis 10 | Bulgaria | – | – | JX256705 | JX256507 | – | JX256605 |
| Sphecodes niger | Czech Republic | x | JX256769 | JX256711 | JX256513 | JX256805 | JX256611 |
| Sphecodes nomioidis 1 | Czech Republic | x | JX256770 | JX256712 | JX256514 | JX256806 | JX256612 |
| Sphecodes nomioidis 3 | Czech Republic | – | – | – | JX256515 | – | JX256613 |
| Sphecodes olivieri | Tunisia | x | – | JX256713 | JX256516 | – | JX256614 |
| Sphecodes pellucidus 1 | Slovakia | x | JX256771 | JX256714 | JX256517 | – | JX256615 |
| Sphecodes pellucidus 11 | Czech Republic | – | – | JX256715 | JX256518 | – | JX256616 |
| Sphecodes pellucidus 3 | Czech Republic | – | – | JX256716 | JX256519 | – | JX256617 |
| Sphecodes pellucidus 4 | Switzerland | – | – | JX256717 | JX256520 | – | JX256618 |
| Sphecodes pellucidus 5 | Hungary | – | – | JX256718 | JX256521 | – | JX256619 |
| Sphecodes pinguiculus | Cape Verde | x | JX256773 | JX256720 | JX256523 | JX256807 | JX256621 |
| Sphecodes pseudofasciatus | Czech Republic | x | JX256772 | JX256719 | JX256522 | – | JX256620 |
| Sphecodes puncticeps 1 | Hungary | x | JX256775 | JX256723 | JX256526 | JX256809 | JX256624 |
| Sphecodes puncticeps 11 | Morocco | – | – | JX256724 | JX256527 | – | JX256625 |
| Sphecodes ranunculi | – | x | – | FJ582493.1i | AF140325.1c | AY227961.1d | AY222597.1e |
| Sphecodes reticulatus 2 | Czech Republic | x | – | JX256726 | JX256529 | JX256811 | JX256627 |
| Sphecodes reticulatus 3 | Italy | – | – | JX256727 | JX256530 | – | JX256628 |
| Sphecodes rubicundus | Czech Republic | x | JX256776 | JX256725 | JX256528 | JX256810 | JX256626 |
| Sphecodes ruficrus 1 | Tunisia | x | JX256778 | JX256728 | JX256531 | JX256812 | JX256629 |
| Sphecodes ruficrus 2 | Spain | – | – | JX256729 | JX256532 | – | JX256630 |
| Sphecodes rufiventris 1 | Czech Republic | x | JX256779 | JX256730 | JX256533 | JX256813 | JX256631 |
| Sphecodes rufiventris 2 | Czech Republic | – | – | JX256731 | JX256534 | – | JX256632 |
| Sphecodes rufiventris 3 | Italy | – | – | JX256732 | JX256535 | – | JX256633 |
| Sphecodes rufiventris 4 | Czech Republic | – | – | JX256733 | JX256536 | – | JX256634 |
| Sphecodes scabricollis 3 | Germany | x | – | – | JX256544 | – | JX256644 |
| Sphecodes scabricollis 4 | Germany | – | JX256781 | JX256742 | JX256545 | JX256815 | JX256645 |
| Sphecodes schenckii 1 | Turkey | x | JX256754 | JX256660 | JX256460 | JX256791 | JX256558 |
| Sphecodes schenckii 2 | Iran | – | – | JX256661 | JX256461 | – | JX256559 |
| Sphecodes sp. 17 | Canada | x | JX256780 | JX256734 | JX256473 | JX256814 | JX256635 |
| Sphecodes sp. 18 | Canada | – | – | JX256735 | – | – | JX256636 |
| Sphecodes sp. 2 | Canada | – | – | JX256736 | JX256538 | – | JX256637 |
| Sphecodes sp. 21 | Japan | – | – | JX256737 | JX256539 | – | JX256638 |
| Sphecodes sp. 26 | – | – | – | AF102844.1b | AF140324.1c | AY227960.1d | AY222596.1e |
| Sphecodes sp. 3 | Canada | – | – | JX256738 | JX256540 | – | JX256640 |
| Sphecodes sp. 7 | USA | – | – | JX256739 | JX256541 | – | JX256641 |
| Sphecodes sp. 8 | USA | – | – | JX256740 | JX256542 | – | JX256642 |
| Sphecodes sp. 9 | USA | – | – | JX256741 | JX256543 | – | JX256643 |
| Sphecodes spinulosus 1 | Czech Republic | x | JX256774 | JX256721 | JX256524 | JX256808 | JX256622 |
| Sphecodes spinulosus 2 | Hungary | – | – | JX256722 | JX256525 | – | JX256623 |
| Sphecodes zangherii 2 | Switzerland | x | – | JX256743 | – | – | JX256646 |
| Sphecodes zangherii 3 | Switzerland | – | – | JX256744 | – | – | JX256647 |
| Systropha curvicornis | – | x | AY654516.1a | – | AF435411.1g | AY227925.1d | AY222562.1e |
| Thrinchostoma lemuriae | – | x | – | – | EU203254.1e | EU203285.1e | EU203223.1e |
| Xeralictus bicuspidariae | – | x | AY654517.1a | – | AF435413.1g | AY227927.1d | AY222564.1e |
At sequences, which are obtained from NCBI database, links to the references are quoted: a Danforth et al. [78], b Danforth [79], c Danforth et al. [80], d Danforth et al. [81], e Danforth et al. [82], f Cardinal and Danforth (unpublished), g Danforth [83], h Danforth et al. [84], i Sheffield et al. [85], j Danforth and Brady (unpublished), k Soucy and Danforth [86], l Danforth et al. [87], m Danforth and Ji [88].
Preparation of DNA sequences
DNA was extracted from part of the abdomen or from the entire individual and then homogenized. DNA was isolated using the Dneasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol. Partial sequences of the following five genes were amplified by PCR: cytochrome oxidase subunit I (COI), elongation factor-1 alpha F2 copy (EF1), wingless (WG), long-wavelength rhodopsin (LWR) and 28S ribosomal RNA (28S). A list of the primers and PCR conditions is given in Table 2. The sequences were edited using the program 4Peaks 1.7.2 [41], then aligned in Clustal W and realigned manually using BioEdit 7.0.9 [42]. All of the alignments are available from www.aculeataresearch.com and www.treebase.org.
Table 2. List of primers including PCR conditions.
| Locus | Primer | Orientation | Sequence 5′→3′ | Reference |
| COI a | ||||
| AP-J-1991 | Forward | TAT AGT TAT ACC ATT TTA ATT G | modified from Pedersen [89] | |
| AP-J-2013 | Forward | GGA GGA TTT GGA AAT TGG CTT ATT CC | modified from Simon et al. [90] | |
| AP-J-2511 | Forward | GAA GTT TAT ATT TTA ATT TTA CCT GG | modified from Simon et al. [90] | |
| AP-N-2536 | Reverse | CCA GGT AAA ATT AAA ATA TAA ACT TC | modified from Simon et al. [90] | |
| AP-N-2950 | Reverse | GCA AAT ACA GCA CTT ATT GA | modified from Pedersen [89] | |
| AP-N-2980 | Reverse | GGA WAT CCA TGA ATA AAT CTT G | unpublished | |
| Ron | Forward | GGA TCA CCT GAT ATA GCA TTC CC | Simon et al. [90] | |
| Pat | Reverse | TCC AAT GCA CTA ATC TGC CAT ATT A | Simon et al. [90] | |
| EF1 b | ||||
| HaF2For1 | Forward | GGG YAA AGG WTC CTT CAA RTA TGC | Danforth et al. [80] | |
| F2-Rev1 | Reverse | A ATC AGC AGC ACC TTT AGG TGG | Danforth et al. [80] | |
| WG c | ||||
| beewgFor | Forward | TGC ACN GTS AAG ACC TGY TGG ATG AG | Danforth et al. [81] | |
| Lep wg2a | Reverse | ACT ICG CAR CAC CAR TGG AAT GTR CA | Danforth et al. [81] | |
| LWR d | ||||
| LWRhFor | Forward | AAT TGC TAT TAY GAR ACN TGG GT | Danforth et al. [81] | |
| LWRhRev | Reverse | ATA TGG AGT CCA NGC CAT RAA CCA | Danforth et al. [81] | |
| 28S e | ||||
| 28SD3for | Forward | GAC CCG TCT TGA AAC ACG GA | Schulmeister [91] | |
| 28SD3rev2 | Reverse | CCC ACA GCG CCA GTT CTG CTT ACC | Schulmeister [91] | |
PCR conditions: 94°C 5 min, 40×(94°C 1 min, 37–50°C 1 min, 72°C 1 min 50 s), 72°C 5 min;
PCR conditions: 94°C 5 min, 35×(94°C 1 min, 54–60°C 1 min, 72°C 1 min 50 s), 72°C 5 min;
PCR conditions: 94°C 5 min, 35×(94°C 45 s, 58°C 45 s, 72°C 45 s), 72°C 5 min;
PCR conditions: 94°C 5 min, 35×(94°C 1 min, 52–60°C 1 min, 72°C 1 min), 72°C 5 min;
PCR conditions: 94°C 5 min, 35×(94°C 45 s, 52°C 45 s, 72°C 45 s), 72°C 5 min.
Phylogenetic analysis
Two concatenated alignments were created using the web application FaBox 1.35 [43]. The main alignment (Align1) contains data from only the reduced dataset (one specimen per species) and sequences of all five genes for a total of 3,679 nucleotide sites (873 base pairs [bp] for COI, 1,152 bp for EF1 including a 394 bp long intron, 403 bp of WG, 632 bp of LWR including a 174 bp long intron and 619 bp of 28S). Align1 was used for the phylogenetic analysis, molecular dating and the reconstruction of ancestral character states. A supplementary alignment (Align2) contains data from the complete dataset but for only three genes (COI, EF1, WG), for a total of 2,428 nucleotide sites. Align2 was used for the supplementary phylogenetic analyses.
Bayesian and maximum likelihood (ML) analyses were performed for each concatenated alignment. Initially, several alignment subsets were created for each gene according to codon position, introns, exons or stem and loops. The best DNA substitution model was chosen for each subset and for the concatenated alignments based on the AICc value (the AIC value [44] corrected for sample size) [45], [46] using the program jModelTest 2 [47]. If the best model was not implemented in the MrBayes program [48], the best implemented model was chosen for Bayesian analysis. Subsequently, several Bayesian and ML analyses were performed with different partitioning schemata for each gene (no partitioning, intron/exon, intron/exon 1st +2nd/3rd exon, intron/exon 1st/2nd exon/3rd exon and stem/loop).
The results of the Bayesian analyses of all partitioning strategies for each gene were compared using Bayes factors [49]. The marginal likelihoods were estimated as the harmonic mean of the likelihood scores using the sump command in MrBayes. The statistic 2 ln(BF) was then calculated as 2[ln(HM1) – ln(HM2)], where HM1 is the harmonic mean of the posterior sample of likelihoods from the one partitioning strategy and HM2 is the harmonic mean of the posterior sample of likelihoods from the other partitioning strategy. According to Brandley et al. [50], values of 2 ln(BF) less than 2 indicate that the first partitioning strategy (HM1) is not significantly better than the second (HM2). Subsequently, the best partitioning strategy for each gene was determined. The results of the maximum likelihood analyses of all the partitioning strategies for each gene were compared using the AIC values [44]. The partitioning strategy with the lowest AIC was considered the best strategy.
Subsequently, three analyses of the concatenated alignments of different partitioning schemata were performed: (1) no partitioning, (2) the most partitioned schema (by the codon positions of each gene and stem and loop structures) and (3) the best partitioning schemata for each gene. The three partitioning strategies of the concatenated alignments were compared using the Bayes factor and the AIC value in the same way as the partitioning schemata of particular genes were compared. Finally, the best partitioning strategies for each method (Bayesian and ML) were determined.
All Bayesian analyses were conducted using MrBayes [48] implemented in BioPortal [51]. Each analysis consisted of running four simultaneous chains for 10 million generations and saving every thousandth tree. Two independent analyses starting from different random trees were performed for each partitioning strategy. The convergence of chains was inspected by checking the posterior distributions of log likelihoods using the program Tracer [52]. The first 25% of trees of each independent analysis were discarded as burn-in. A consensus tree was created from the best partitioned analyses according to the Bayes factor.
All of the ML analyses were calculated using the program Garli [53] implemented in BioPortal [51]. Four independent search replicates were performed for each analysis. One thousand bootstrap replicates were performed for calculating the branch support values of the best partitioned strategy (according to the AIC value) for Align1 and Align2. A bootstrap consensus tree was constructed with the program PAUP 4.0b10 [54].
Definition of character states for character mapping and test of irreversible evolution
Two types of characters were mapped onto the phylogenetic tree of the genus Sphecodes: the ancestral host group and the ancestral host specificity (specialist or generalist). The list of 37 Sphecodes species with mapped characters is given in Table 3.
Table 3. List of mapped characters.
| Taxon | Specialization | Hosts | |||||||
| LS | LO | H | S | A | C | P | M | ||
| S. albilabris (Fabricius) | (S) G (G) | x | x | x | |||||
| S. alternatus Smith | S | x | |||||||
| S. autumnalis Mitchella, b | S | x | |||||||
| S. clematidis Robertson | ? | ||||||||
| S. confertus Sayc | S | x | |||||||
| S. crassus Thomson | (S) G (G) | x | |||||||
| S. cristatus Hagens | (S) S (G) | x | x | ||||||
| S. croaticus Meyer | S | x | |||||||
| S. dusmeti Blüthgen | ? | ||||||||
| S. ephippius (Linnaeus) | G | x | x | x | x | x | |||
| S. ferruginatus Hagens | S | x | |||||||
| S. geoffrellus (Kirby) | (S) G (G) | x | |||||||
| S. gibbus (Linnaeus) | (S) G (G) | x | x | ||||||
| S. hyalinatus Hagens | S | x | |||||||
| S. longuloides Blüthgend | S | x | |||||||
| S. longulus Hagens | S | x | |||||||
| S. majalis Pérez | S | x | |||||||
| S. marginatus Hagens | S | x | |||||||
| S. miniatus Hagens | S | x | |||||||
| S. monilicornis (Kirby) | G | x | x | x | x | x | |||
| S. niger Hagens | S | x | |||||||
| S. nomioidis Pesenko | ? | ||||||||
| S. olivieri Lepeletier | S | x | x | ||||||
| S. pellucidus Smith | (S) G (G) | x | x | ||||||
| S. pinguiculus Pérez | S | x | |||||||
| S. pseudofasciatus Blüthgen | ? | ||||||||
| S. puncticeps Thomson | S | x | |||||||
| S. ranunculi Robertson | ? | ||||||||
| S. reticulatus Thomson | (S) G (G) | x | x | ||||||
| S. rubicundus Hagens | S | x | |||||||
| S. ruficrus (Erichson) | S | x | |||||||
| S. rufiventris (Panzer) | S | x | |||||||
| S. scabricollis Wesmael | S | x | |||||||
| S. schenckii Hagens | S | x | |||||||
| S. sp. 17 | ? | ||||||||
| S. spinulosus Hagens | S | x | |||||||
| S. zangheri Noskiewicz | ? | ||||||||
The specializations are S, specialist and G, generalist. The state used in the main analysis (Distribution II) is shown in bold, that used in Distribution I is shown on the left in parentheses, and that used in Distribution III is shown on the right. The hosts are LS, Lasioglossum sensu stricto; LO, Lasioglossum other; H, Halictus subgenus Halictus; S, Halictus subgenus Seladonia; A, Andrena; C, Colletes; P, Perdita; and M, Melitturga. If unspecified, information on specialization and hosts is adopted from Bogusch and Straka [39].
Ancestral host group
The list of known host species of Sphecodes used in our analyses was adopted from data in Bogusch and Straka [39], Michener [40], Blüthgen [55], Danforth [56] and Eickwort [57]. The hosts of Sphecodes species were divided into several groups according to their genera (Andrena, Colletes, Perdita, and Melliturga) and used to map the ancestral hosts. Exceptions to this practice were the most frequent hosts of the genera Lasioglossum and Halictus, which were each divided into two main lineages according to their phylogeny (Lasioglossum sensu stricto and other Lasioglossum lineages and Halictus subgenus Halictus and Halictus subgenus Seladonia lineages) [58].
Ancestral host specificity
We classified all Sphecodes species as either specialists or generalists according to their known host range(s) [55], [56], [57], [39]. However, there are no agreed-upon definitions of specialists and generalists, and moreover, the two categories are continuous rather than discrete. Therefore, the fundamental question is the position of the dividing line between the specialists and the generalists. Several criteria can be used to estimate the degree of resource specialization. The most commonly used criterion is the number of resources utilized by a species. Other criteria that can be considered are the differences in the intensity of use of particular resources [59], the degree of phylogenetic distinction between the utilized resources [60], the ecological distinction between the utilized resources and the number of adaptations necessary for successful resource use. There are several indices available that measure the degree of host specificity in parasites [61]; however, such indices require detailed knowledge concerning the individual species, which is not available for Sphecodes. Moreover, none of these indices account for ecological differences between the utilized resources. Therefore, we did not rely on any existing index; instead, we defined specialists and generalists as a function of the phylogenetic and ecological distinctions between the utilized resources. To validate our approach, we used the following three alternative definitions and thus three alternative distributions of specialists and generalists.
Distribution I – the most unrestrictive definition of specialists
1) Specialists comprise species whose hosts represent a single genus or include one additional host of a different genus, but the second host is only sporadically used. 2) Generalists include species that do not meet either of the above criteria for specialists.
Distribution II – used for our main analysis
1) Specialists comprise species with ecologically similar hosts of the same genus or with one additional host of a different genus that is nonetheless ecologically similar to the other hosts and only sporadically utilized. 2) Generalist species do not meet either of the above criteria for specialists.
Distribution III – the most restrictive definition of specialists
1) Specialists comprise species with ecologically similar and related hosts that represent a single genus. 2) Generalist species do not meet this criterion for specialists.
Reconstruction of ancestral states
Each terminal in the Sphecodes phylogenetic tree was coded by character state, that is, either by host type (Lasioglossum s. s., other Lasioglossum, Halictus subgenus Seladonia, Halictus subgenus Halictus, Andrena, Colletes, Perdita and Melliturga) for analyses of the reconstruction of the ancestral host group or by behavioral category (specialist and generalist) for analyses of the reconstruction of ancestral host specificity. For the species with no known host, the terminal was coded as “missing data”. Two methods for reconstructing the ancestral states were used: the Bayesian method using the program BayesTraits [62] and the maximum parsimony method using the program Mesquite [63].
Bayesian method
A tree file was created from the Bayesian analysis of the phylogeny. Each 10,000th tree (each 10th saved tree) of each independent run was selected except for the first 25% of trees, which were discarded as burn-in. Therefore, the complete tree file consisted of 1,500 trees. Three alternative data files (according to the three alternative distributions of specialists and generalists) for the reconstruction of ancestral host specificity were created. Ancestral states for all the nodes were calculated for Distribution II. Only the ancestral states of Node 1 (i.e., the most recent common ancestor of the genus Sphecodes) were calculated for Distributions I and III.
An independent analysis for each host type was performed in the mapping of the ancestral hosts. Thus, a data file for each host was created with both the presence and the absence of the host in each Sphecodes species. We chose this method instead of using only one analysis with multiple character states to assign independent posterior probabilities for several different hosts at one ancestral node. This model reflects independent transitions between the host genera in cuckoo bee evolution, unlike the standard multiple-character analysis in which the posterior probability is distributed within all the states and provides an inconclusive result concerning the number of possible host genera at the ancestral nodes. Multiple-character analysis of all the genera together can well resolve which host genus was more probable at which ancestral node, but this is not the question here. The use of the generic level for hosts, however, underestimates the host switches but better reflects the current knowledge of the host associations included in the model. Studied host switches quite likely also exist on the subgeneric level, but these switches are missed in our study.
We initially performed the ML analyses for each set of characters (hosts, Distribution I, Distribution II and Distribution III). These analyses computed the optimal transition rates from one character state to another for each tree in the tree file. The settings of the Bayesian analyses were subsequently optimized to best reflect the distribution of the transition rates obtained from the ML analyses.
A reversible jump model with a hyperprior was used for all the analyses. The reversible jump model searches among the possible models of trait evolution (those with the same versus different transition rates between character states) and visits these models in proportion to their posterior probabilities [64]. Hyperprior means that the program estimates priors from the data using a uniform hyperprior to seed the priors [62].
The priors were obtained from an exponential distribution in the interval 0–20 for the reconstruction of the ancestral host group, 50–200 for the reconstruction of the ancestral host specificity using Distributions II and III, or 0–70 for the reconstruction of ancestral host specificity using Distribution I.
The ratedev parameter, which specifies the magnitude of change proposed to rate the coefficients at each iteration of the chain, was optimized for the resulting acceptance rate of the newly proposed values of the rate parameters between 20–40%, as is recommended in the manual [62].
The analysis was run for either 5 million (the reconstruction of ancestral hosts) or 50 million (the reconstruction of ancestral host specificity) generations. The estimated parameters were checked for convergence using Tracer 1.4 [52]. The first 2 million (the reconstruction of ancestral host) or 40 million (the reconstruction of ancestral host specificity) generations were discarded as burn-in.
The mean of the posterior probabilities of the character state was calculated for each node. Only hosts with a posterior probability greater than 0.7 were considered as possible hosts.
To test whether there is significant support for a character state with a higher posterior probability, an MCMC analysis with a fixed character state as a specialist and alternatively as a generalist was performed for 50 million generations for each node. The harmonic means resolved from these two analyses were compared using the Bayes factor (see above). Values above 2 are considered to indicate positive support, those above 6 are considered to indicate strong support, and those above 10 are considered extremely strong support [50].
Maximum parsimony method
A consensus tree generated from the Bayesian phylogenetic analysis was used for the maximum parsimony ancestral mapping method. The ancestral host reconstruction and the three alternative analyses of ancestral host specificity reconstruction corresponding to the three alternative distributions of specialists and generalists were performed using the program Mesquite [63].
Test of irreversible evolution
We tested whether evolution is directional, i.e., whether specialists can arise from generalists, but generalists cannot arise from specialists (or vice versa), or flexible, i.e., specialists can arise from generalists, and vice versa. Therefore, a model allowing transitions in both directions was compared to a model allowing transitions in only one direction. Likelihoods for each tree in the tree file (the same tree file as that used in the character mapping analyses) were calculated using the program BayesTraits for (i) a model allowing transitions in both directions, (ii) a model allowing transitions only from generalist to specialist and (iii) a model allowing transitions only from specialist to generalist. The mean of the likelihoods was calculated for each model, and the model means were compared using the likelihood ratio test. The two types of model differ by one free parameter, and the resulting value approximates a chi-square distribution with one degree of freedom [62].
Results
Phylogeny of subtribe Sphecodina
Both phylogenetic analyses (Align1 and Align2) were performed without dataset partitioning. We applied the DNA substitution model, GTR + Γ + I, to the entire dataset. This partitioning strategy is suitable for the ML method (for Align1, AIC [no partitioning] = 64,992, AIC [best partitioning schema for each gene] = 65,048, and AIC [the most partitioned schema] = 65,078; for Align2, AIC [no partitioning] = 53,240, AIC [best partitioning schema for each gene] = 53,246, and AIC [the most partitioned schema] = 53,260) as well as for the Bayesian method, where all the comparisons of 2 ln (BF) between models were <2.
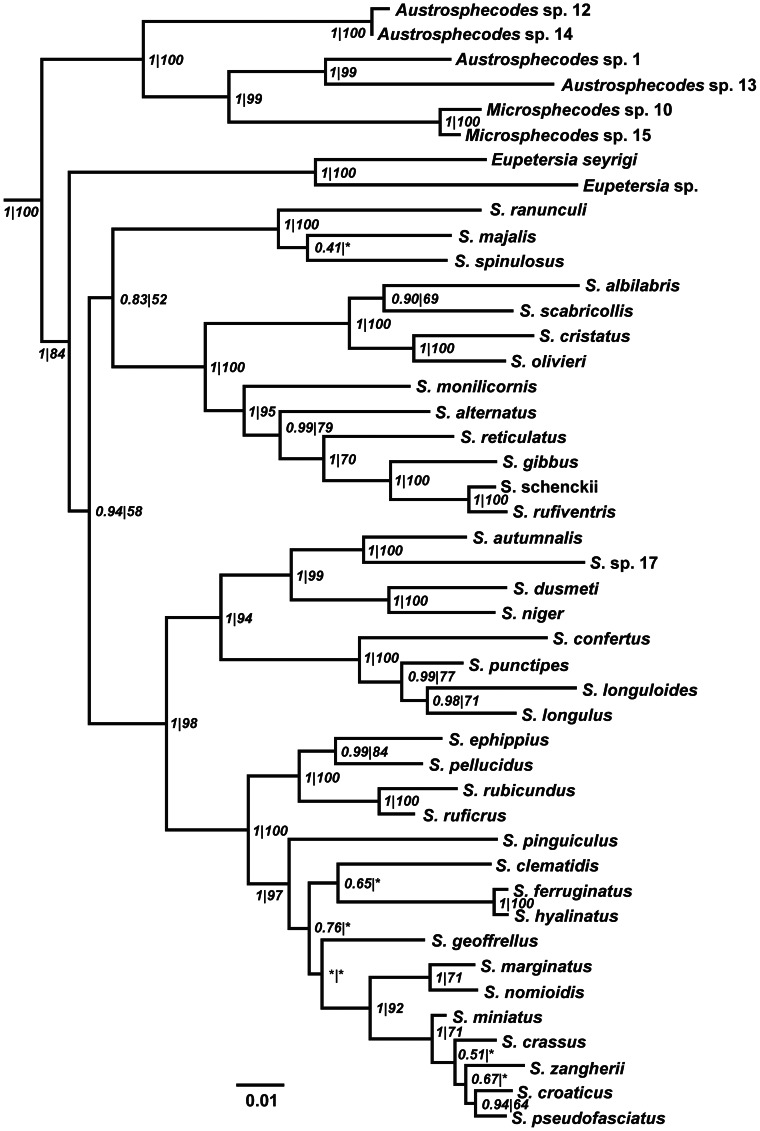
All the phylogenetic analyses (the Bayesian and ML methods for Align1 and Align2) resulted in robust phylogenies, all of which were congruent at the majority of nodes (Figs. 1 and S1, all trees are available from www.treebase.org). There is some difference between the tree topologies constructed from the five- and the three-gene analyses; however, branch support increases with the addition of sequential information at the majority of nodes. The main difference between the resulting trees concerns the ambiguity of branching of the basal lineages of the subtribe Sphecodina. Both of the analytical methods used for branch support (posterior probability in the Bayesian and bootstrap in the ML analysis) give good support at most nodes for Align1 (Figs. 1 and S1). The Bayesian tree with topologies based on the analysis of Align2 with posterior probabilities and the ML bootstrap support are available in the Supporting Information (Fig. S1).
Figure 1. Phylogenetic tree resulting from Bayesian analysis of Align1.
Posterior probabilities are in front of the slash and bootstrap values from the maximum likelihood analysis are behind the slash. Posterior probabilities below 0.5 and bootstrap values below 50 are replaced by an asterisk (*).
Two taxa used in our study from the Microsphecodes genus cluster within four specimens of Austrosphecodes, which renders Austrosphecodes a strongly supported paraphyletic group (Fig. 1).
The consensus tree from the Bayesian analysis, along with the ML tree, suggests the monophyly of Sphecodes but with weak statistical support (Align1 posterior probability 0.94, bootstrap 58; Align2 posterior probability 0.83, bootstrap 51). It is unclear whether the genus Eupetersia forms a sister group of genus Sphecodes or belongs within it.
The position of the clade consisting of S. majalis, S. spinulosus and S. ranunculi (Proteraner group) is uncertain because its support within the genus Sphecodes was very weak (Align1 posterior probability 0.83, bootstrap 52; Align2 posterior probability 0.53) and even absent based on the ML analysis of Align2 in which this Proteraner group appears as a sister group to all the other Sphecodes species. Some other Sphecodes species were problematic in our analysis, e.g., S. pinguiculus, S. geoffrellus and the entire S. marginatus group (Figs. 1 and S1).
All of the specimens of the morphologically and behaviorally diversified species known to be individual specialists (S. ephippius and S. monilicornis) are well supported as members of a monophyletic group and do not vary more than specialized species in branch lengths (Fig. S1).
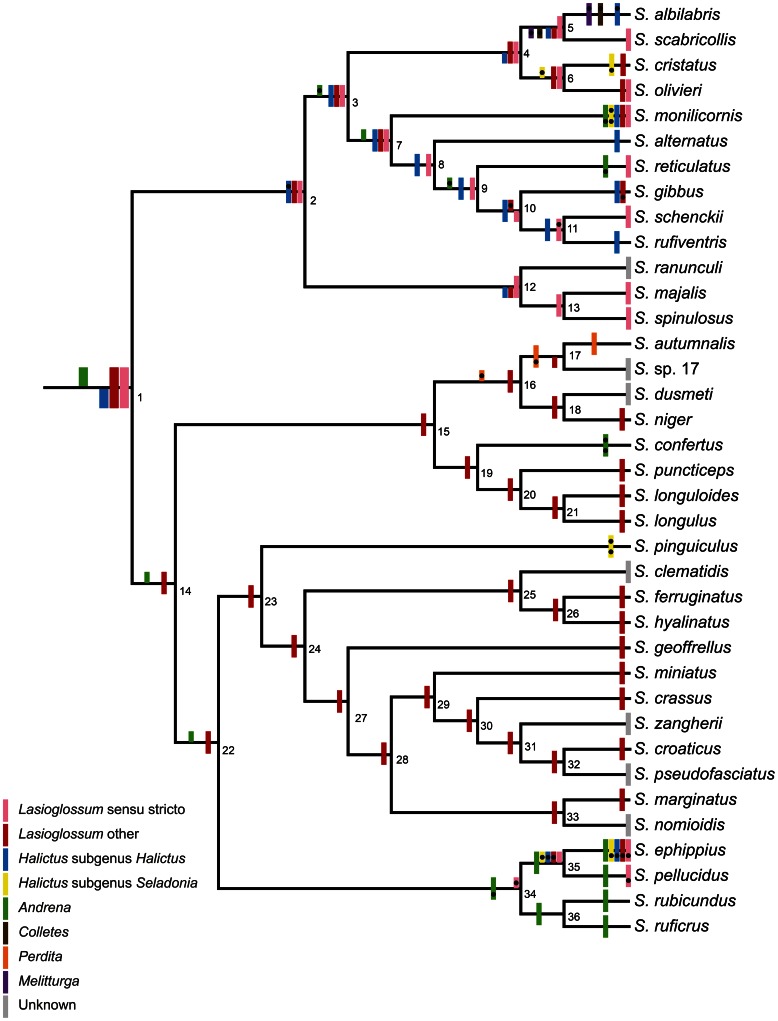
Ancestral host group
Both of the ancestral mapping methods (Bayesian and maximum parsimony) identified the ancestral Sphecodes s.l. as having three host lineages: Lasioglossum s. s., other Lasioglossum and either Andrena (according to the Bayesian method) or Halictus (according to the maximum parsimony method) (Fig. 2, Table S2). Both analyses showed that switches between hosts have commonly occurred (Fig. 2). These switches have occurred 17 times at 13 ancestral nodes of Sphecode according to the Bayesian analysis and 16 times at 10 ancestral nodes according to the maximum parsimony analysis (Fig. 2). Only three recognized switches were the same in both analyses: S. monilicornis (switching to Halictus subgenus Seladonia within its five host groups), S. confertus and S. pinguiculus. In ten cases, it is clear that the switches correspond to one another, although they occur at different locations in each analysis (the localization of the host group according to the Bayesian analysis precedes the dash): Node 5– S. albilabris (switches to Colletes and Melitturga), Node 6– S. cristatus (switch to Halictus subgenus Seladonia), Node 9– S. reticulatus (switch to Andrena), Node 10– S. gibbus (switch to Lasioglossum), Node 16– Node 17 (switch to Perdita), Node 34– S. ephippius and S. pellucidus (switch to Lasioglossum s. s.), and Node 35– S. ephippius (switch to Lasioglossum other, Halictus subgenera Halictus and Seladonia). In four cases, a switch is apparent from the Bayesian analysis but not the maximum parsimony analysis: Node 2 (switch to Halictus), Node 3 (switch to Andrena), S. albilabris (switch to Halictus) and Node 11 (switch to Lasioglossum s. s.). In one case, a switch apparent from the maximum parsimony analysis is absent from the results of the Bayesian analysis: Node 34 (switch to Andrena). A complete list of the posterior probabilities of the particular ancestral states is given in Table S2.
Figure 2. Character state reconstruction of ancestral hosts of the cuckoo bees of the genus Sphecodes .
The possible hosts identified by Bayesian analysis (posterior probability >0.7) are shown above the nodes and those identified by maximum parsimony analysis are shown below the nodes. Black spots indicate host switches. Specific values of the posterior probabilities are shown in Table S2.
Evolution of host specificity
We performed host specificity analyses using three alternative definitions of specialist and generalist, which have different character distributions in the phylogeny of Sphecodes. Based on our definitions of host specificity, we identified 2, 8 and 9 host generalists (using Distributions I, II and III, respectively) from 30 species with known host records and 6 distinct species with unknown host records. Other Sphecodes specimens belonging to taxa with unknown host records as well as unidentified specimens were not included in the analysis. According to the likelihood ratio test and using Distribution II or III, the bidirectional model for the evolution of host specificity (character states can change in either direction between specialist and generalist) has a significantly higher likelihood than any unidirectional model. The likelihood ratio test using Distribution I indicates that the bidirectional model is significantly better than the unidirectional model for change from generalist to specialist, but the unidirectional model for change from specialist to generalist is not significantly worse than the bidirectional model (Table 4).
Table 4. Results of likelihood ratio test.
| Distribution I | |||
| G ↔ S | G → S | S → G | |
| Mean of ln likelihood | −6.545 | −8.731 | −7.850 |
| Result of LR test | 4.370 | 2.610 | |
| P – value | 0.0365 | 0.1061 | |
Comparison of results from one-direction models of character evolution with an unrestricted model for specialists (S) and generalists (G) for three definition criteria of host specialization (Distributions I-III).
The results of the host specificity mapping are largely ambiguous. According to the Bayesian ancestral state reconstructions, the specialist state is the more probable host specificity at the majority of nodes, including the most recent common ancestor of Sphecodes (Table S1, Fig. S2). However, these results are mostly inconclusive. The Bayes factor positively supports particular ancestral states only in 4 cases (specialists at nodes 11, 15, 26, and 36) (Table S1, Fig. S2). The BF value is less than 2 (indicating no support) for the remaining nodes. These results indicate that generalists and specialists are approximately equally probable for the majority of nodes, and we cannot determine whether the ancestors of certain extant species were specialists or generalists.
According to the maximum parsimony analyses, the generalist state occurs only at Node 35 using Distribution III. Both states are recognized as possible at Nodes 7, 9, and 10 using Distribution II and at Nodes 3, 4, 5, 6, 7, 9, and 10 using Distribution III. For the remaining nodes, the specialist state is the more probable (Fig. S2). According to the results of the maximum parsimony method, the most recent common ancestor of Sphecodes was a specialist, and generalist species must have arisen from specialist ancestors only recently.
Discussion
Phylogeny and systematics
Our results support the monophyly of the subtribe Sphecodina in accordance with previous assumptions [40], [24] but conflict with other suggested relationships within this subtribe (Fig. 1, see also Fig. S1). Michener [40], [24] recognizes Austrosphecodes as a subgenus of Sphecodes and Microsphecodes as a separate genus from Sphecodes. However, our results do not support this classificatory hypothesis. Our analyses suggest that Austrosphecodes is not closely related to Sphecodes but is a basal lineage of Sphecodina, and Microsphecodes is an inner group of Austrosphecodes, thus rendering the latter paraphyletic (Fig. 1). Ptiolocleptis (missing in our study) may also be a representative of this lineage [40]. Little is known about the diversity and species relationships within this Neotropical lineage, and thus, other taxonomic suggestions would not currently be useful (e.g., dividing all lineages into different genera or subgenera). The genera Eupetersia and Sphecodes are recognized as monophyletic by Michener [24]; however, clade support for this supposition was relatively weak in our analysis, although we omit certain taxa that may affect inference of the basal relationships of this subtribe (Callosphecodes Friese and Nesosphecodes Engel) [65], [66]. Further studies are necessary to resolve the basal relationships within the subtribe Sphecodina and the systematics of Sphecodes at the subgeneric level. The groups established by Robertson [67] based on morphological descriptions, such as Proteraner (here, S. majalis, S. ranunculi and S. spinulosus) or Drepanium (here, S. confertus, S. longuloides, S. longulus and S. punctipes) seem plausible. Our phylogenetic analysis and the preexisting keys (based on morphology [39]) sort species into the same groups across studies, which may enable researchers to understand characters in this problematic diversified group and permit the further distinction of Sphecodes species to the level of subgenera. Such taxonomic studies will enhance detailed alpha-taxonomic, morphological and behavioral studies in the future.
Evolution of host specificity
Sphecodes cuckoo bees appear to be largely specialists regarding host choices. Only two of the thirty analyzed species are obvious generalists, although an additional seven exhibit host variability [39]. We mapped host specialization strategies and reconstructed ancestral strategies in Sphecodes cuckoo bees using two different methods (Bayesian and maximum parsimony), but the results of the two methods are incongruent. The results of the Bayesian analysis are largely inconclusive; therefore, the likelihood ratio test of the host specialization evolutionary model seems to be the more relevant approach for answering the question about evolution of host specialization. The likelihood ratio test using Distribution II and III strongly supports the occurrence of clear transitions between generalists and specialists in both directions in the evolution of the genus Sphecodes. Unidirectional evolution from generalists to specialists or from specialists to generalists is significantly rejected. The results of the likelihood ratio test using Distribution I are not so convincing, but the transition from generalists to specialists is also significantly rejected (Table 4). Although the generalist state is traditionally considered to be the ancestral state and to later lead to the derived condition (specialization) [7], here we provide evidence of both types of evolutionary change. There are two possible reasons for the reversal to the ancestral state and back.
First, perhaps the generalist state is not the ancestral one. According to Emery [68], social ant parasites arise from their hosts as a result of sympatric speciation and must therefore be specialists at the outset. Nevertheless, environmental changes apply selective pressures to all parasitic species, and sometimes, adaptation to this situation requires the adoption of new or additional host species to avoid extinction. If such host switches are successful, parasitic species may become generalists (at least for a short time). Several authors adopted this theory, recently interpreted as Emery's rule, e.g., Carpenter et al. [69], who applied the theory to explain the specialization of cuckoo wasps to their closely related hosts. Emery's rule has been used for social parasites, but we do not know if it applies to nest cleptoparasites.
Second, Sphecodes cuckoo bees may not have reverted to the generalist strategy as understood for nonspecialized ancestors, but may have adopted a new generalist strategy of specialization at the individual level. This strategy occurs in two European Sphecodes species (S. monilicornis and S. ephippius), which have the broadest host range [38]. Specialization at the individual level allows species to take advantage of a large number of hosts as well as potentially adapt to a particular host or at least overcome certain constraints. Such specialization might be coded as a third type of host strategy, along with specialist and generalist.
Third, generalist species can consist of distinct genetically separated lineages, each specialized to a different host. However, no clustering within lineages is evident for S. ephippius or S. monilicornis in our phylogenetic trees based on all of the samples (Fig. S1), albeit that the genes chosen for our study could be inappropriate for resolution of relationships among such close lineages.
Our results based on the mapping of host specificity and the likelihood ratio test are likely biased by the problematic determination of the basic distinction between the specialist and generalist strategies. There is also the limitation that the mapping of ancestral host specificity cannot detect a host switch that occurs without a change in host specificity strategy. For these reasons, the character state reconstruction of the ancestral host group of Sphecodes cuckoo bees offers a better approach and, unlike the mapping of host specificity, provided comparable results using both applied methods (the Bayesian and maximum parsimony analyses). Practically, this reconstruction provides a different approach to the same problem discussed above and can also determine the number of possible host switches and the host lineages that become new hosts. Despite obvious limits on lineage resolution, we can identify the most prominent host changes. There are between 11 and 13 clades with 16 to 17 detected host changes (depending on analysis method) in the evolutionary history of the analyzed Sphecodes species (Fig. 2). Although we missed all the host switches below the resolution of our host definitions, our results suggest that cuckoo bees are relatively flexible in their host choice. Thus, there is a high probability that our finding of multiple switches between the generalist and specialist strategies in cuckoo bee ancestors is valid.
We obtain a similar result from the analysis of the most recent common ancestor of all the Sphecodes species. It is difficult to infer whether this ancestor was a specialist or a generalist, but it seems to have been a parasite of two or possibly three different lineages (Figs. 2 and S2). However, a pitfall of the ancestral host group reconstruction method arises because some identified hosts might not have yet existed when the ancestral Sphecodes species lived and diverged. According to Gibbs et al. [58], [70], the most recent common ancestor of Sphecodes lived before approximately 25 MYA, and the divergence of host Lasioglossum lineages (Lasioglossum s. s. and other subgenera of this genus) occurred before approximately 30 MYA. These results suggest that the Lasioglossum lineages had already diverged when the MRCA of Sphecodes arose. However, the credible dating intervals are quite large and overlap each other (18–4 MYA for the MRCA of Sphecodes and 25–7 MYA for divergence of the Lasioglossum lineages). Therefore, cospeciation of the Sphecodes ancestor with the Lasioglossum ancestor is also a possible scenario.
Although Lasioglossum is strongly supported as an ancestral host lineage, hosts from Andrena or the Halictus lineage are also likely for the most recent common ancestor of Sphecodes. These host groups were already established when the ancestral Sphecodes species lived and diverged [71].
Host switches and constraints for the generalist strategy
According to our results, specialization is not an evolutionary dead end in cuckoo bees of the genus Sphecodes, even though the majority of extant species are specialists (Figs. 2 and S2). Our results clearly suggest that the specialists do not depend entirely on their hosts but likely have a range of potential host species (or lineages) that might not be optimal but are sufficient for survival. According to Agosta and Klemens [22], such a potential host pool may provide a sloppy fitness space (i.e., a potential fitness space outside the range of conditions in which the species evolved). Such a fitness space may enable a switch to another host species. Nevertheless, we could obtain the same result if a large number of extinct species existed and they were all strictly dependent on their hosts (i.e., unable to switch to alternate hosts). Knowledge of the extinction rate of these cuckoo bees would be informative here; however, no fossil cuckoo bees are known (M. S. Engel, personal comm.). We prefer the former hypothesis because the behavior of some recent Sphecodes species suggests the successful utilization of different hosts following exposure to ecological pressure. For example, S. albilabris uses the early spring bee, Colletes cunicularius, as the primary and most likely essential host [55], [72], but this parasite lives longer than its spring host and utilizes various species similar in size to C. cunicularius during the summer [39]. These hosts (e.g., Meliturga clavicornis [73] and Halictus quadricinctus [39]) are phylogenetically unrelated to the main host species and to each other. As our results show, cuckoo bees are most likely not obligately fixed to their hosts; therefore, host switches are more likely than cospeciation events. Nevertheless, there must be certain constraints that inhibit the arbitrary switching of hosts. If so, the most advantageous strategy would be generalism. Three constraints that may act synergistically are the most probable:
Pollen specialization of host species. Pollen-collecting bee species are physiologically constrained in their choice of flowers due to pollen toxicity [25], [26], and it is likely that the host choices of cuckoo bees are physiologically constrained as well. Appropriate hosts are only those that provide pollen appropriate for the growth of the immature cuckoo stages.
Neurological constraints. Bees have limited memory and learning capacities [28], [4]. The limitations of parasitic bees may be similar to the constraints involved in flower constancy [32]. There are likely several different factors involved in finding and using an appropriate host, requiring various olfactory, visual and strategic abilities. The brains of bees can concentrate only on a single target at a time and can be effective only in such cases.
Size ratio of a cuckoo bee to its host. A host smaller than the size of a cuckoo bee would likely be unsuitable. First, the amount of pollen provided may be insufficient for the development of a larva of a large cuckoo bee. This constraint may not be overwhelming because there is considerable variability in the size of generalists (S. monilicornis as well as S. ephippius) (Bogusch and Straka, personal observation). A more important consideration may be the nest entrance size because adult females must be able to enter the host nests. Adult female S. crassus that had most likely grown up in cells designated for queens of the social species, Lasioglossum pauxillum, were unable to penetrate the nests of the same host species after reduction of the entrance size to fit smaller workers of L. pauxillum (J. Straka, personal observation).
Cuckoo bees face one or more of these constraints when switching to ecologically dissimilar hosts. However, these constraining factors need not be limiting if the switch occurs within ecologically similar hosts. Therefore, there is a large difference between a “true generalist” with many ecologically dissimilar and unrelated hosts and a “faux generalist”, which is specialized on only a phylogenetically diversified lineage of hosts [74]. Moreover, there could also be “faux specialists”, which are able to utilize more ecologically dissimilar hosts but remain restricted only to a few ecologically similar resources while the others are hidden (for example due to competition or nonoverlapping distributional ranges) [74]. Therefore, the host specificity of a particular species should be determined by its ability to overcome the above-mentioned constraints. Other methods do not permit the detection of false specialists and generalists.
True generalist species often use dissimilar hosts from various families [38]. How do they overcome these constraints? One possibility is specialization at the individual level, which reduces intraspecific competition and enhances host utilization by individuals. Such generalist species can accept almost any host, even when they are eusocial. Both S. ephippius and S. monilicornis invade the nests of both solitary and eusocial bee species [39].
We can also mention that there are many pollen-collecting bee species that are not attacked by Sphecodes, some that are even closely related to the host species. Such species might be inappropriate for the parasite for several reasons different from the already mentioned constraints. These species might be shifted in time, biotope or biogeography and do not meet the parasite; they might have an effective protective behavior against the parasite evolved during past contact with Sphecodes; or they might display protective behavior against another extant cuckoo parasite from a different lineage, which protects them from effective utilization by Sphecodes. The latter behavior can involve nest construction habits, such as closure of the nest entrance or the digging of a fake nest entrance [75].
Summary of observed behavioral patterns and individual choice constancy
We can find several different life strategies in Sphecodes cuckoo bees. Specialization at the individual level such as that currently known in two unrelated generalist species (S. monilicornis and S. ephippius) seems to occur only rarely. Other species are usually connected to a single host species (or genus) at one locality and time. Nevertheless, at different localities or times, one can find the same specialized species or even the same individuals parasitizing different hosts unrelated to the previously known hosts. Alternative hosts are well described in Sphecodes albilabris [39], [73], and such parasitizing flexibility may also be true for S. cristatus, S. olivieri, S. reticulatus, S. geoffrellus, S. crassus, S. pellucidus, and certain other species that are expected to be specialists but also have different unexpected hosts proposed for them [39]. So-called generalists that are actually specialists at the individual level solved the problem of how to exploit a potentially rich host pool, but other species, even the specialized ones may be able to switch hosts under certain circumstances. Given our results, we can suggest that a flexible host utilization approach should be widespread or even general for cuckoo bees, and thus, the host utilization strategy should be interpreted more broadly than simple generalist and specialist. We have shown that this differentiation into specialists and generalists hardly works at all. Therefore, we looked for analogous patterns of flexible host utilization and for frequent host switches in the evolution of bees, and we found similarities between the choice of host in cuckoo bees and the choice of flower in pollen-collecting bees (flower constancy) [32], [38]. We can imagine host choice as a specialization at the individual level in all species, with individual variability in the decision strategy to switch hosts. Of course, individuals of some species are more constant in their host choice and change the host less frequently (specialists) than others (generalists). However, the degree of flexibility in the host choice of species (as well as individuals) falls more on a continuum than in discrete categories (specialist and generalist). Moreover, various ecological pressures could limit the availability of the preferred host at a given time and therefore the willingness of a single individual (as well as the whole population sharing the same constraints) to switch hosts can vary with time. For example, when an individual of a specialized species has many available oocytes remaining in its ovaries and is unsuccessful in finding an appropriate host, the decision to switch hosts may be made more readily and thus quite frequently. This decision pattern derives from the flower constancy example and can also likely be applied to other choices by bees. The pattern may well represent a common behavioral strategy for decision making to overcome various constraints in bees and other Hymenoptera; we refer to the pattern as “individual choice constancy”. This hypothesis must be tested in the future, but observations from Sphecodes bees are consistent with this view, including the results of laboratory studies, particularly those performed with S. pellucidus [76]. This is the only well-documented case of host switching (from Andrena host to Lasioglossum) in a potentially specialist species [39]. Conducting similar, more detailed experiments with choice pressures in a highly specialized species would be informative. There are also interesting field observations concerning the existence of host “races” in cuckoo bees that differ in size, e.g., in the genus Coelioxys [77]. These observations also suggest that individual specialization is modulated by the constraints proposed in this paper. Experiments varying the size of the cuckoo bee, the pollen source and the configuration of host nests and their entrances would be useful for understanding the existence and evolutionary importance of individual choice constancy in cuckoo bees.
Supporting Information
Phylogenetic tree resulting from Bayesian analysis of a complete dataset (Align2). Posterior probabilities are in front of the slash; bootstrap values of the maximum likelihood analysis are behind the slash. Posterior probabilities lower than 0.5 and bootstrap values lower than 50 are replaced by an asterisk (*).
(TIF)
Character state reconstruction of host specificity in cuckoo bees of the genus Sphecodes . The larger pie charts show posterior probabilities obtained by Bayesian analysis using Distribution II (main analysis). The smaller pie charts show the results (posterior probabilities in Bayesian analyses or possible character states in maximum parsimony analyses) of other analyses in which interpretations differed from those of the main analysis. B1, Bayesian analysis using Distribution I; MP1, maximum parsimony analysis using Distribution I; B3, Bayesian analysis using Distribution III; and MP3, maximum parsimony analysis using Distribution III.
(TIF)
Complete results of character state reconstruction of host specifity. DI: Distribution of specialist and generalist I, DIII: Distribution of specialist and generalist III, P(G): posterior probability for generalism as ancestral state, P(S): posterior probability for specialism as ancestral state, BF: Bayes Factor (support of more probable state). Values of BF lower than 2 are replaced by asterisk (*).
(DOC)
Complete results of character state reconstruction of ancestral hosts. P (LS): posterior probability for Lasioglossum s. s. as ancestral host, P (LO): posterior probability for Lasioglossum other as ancestral host, P (H): posterior probability for Halictus subgenus Halictus as ancestral host, P (S): posterior probability for Halictus subgenus Seladonia as ancestral host, P (A) posterior probability for Andrena as ancestral host, P (C): posterior probability for Colletes as ancestral host, P (P): posterior probability for Perdita as ancestral host, P (M): posterior probability for Melitturga as ancestral host. The values higher than 0.7 are in bold.
(DOC)
Acknowledgments
We would like to thank the colleagues who helped us by providing material for this study, Victor H. Gonzalez along with several anonymous reviewers for valuable contributions to this manuscript and Mike Arduser for important information about Nearctic Sphecodes hosts.
Funding Statement
This project was supported by grant from Czech Science Foundation (www.gacr.cz/), project no. 206/09/0522 and SVV poject Integrative Animal Biology no. SVV-2012-265206 from Ministry of Education, youth and sports. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Van Valen L (1973) A new evolutionary law. Evol Theory 1: 1–30. [Google Scholar]
- 2. Dawkins R, Krebs JR (1979) Arms Races between and within Species. Proc R Soc Lond B Biol Sci 205: 489–511. [DOI] [PubMed] [Google Scholar]
- 3. Futuyma DJ, Moreno G (1988) The evolution of ecological specialization. Annu Rev Ecol Syst 19: 207–233. [Google Scholar]
- 4. Bernays EA (2001) Neural limitations in phytophagous insects: Implications for diet breadth and evolution of host affiliation. Annu Rev Entomol 46: 703–727. [DOI] [PubMed] [Google Scholar]
- 5. Jaenike J (1990) Host specialization in phytophagous Insects. Annu Rev Ecol Syst 21: 243–273. [Google Scholar]
- 6. Whitlock MC (1996) The red queen beats the jack-of-all-trades: The limitations on the evolution of phenotypic plasticity and niche breadth. Am Nat 148: S65–S77. [Google Scholar]
- 7. Kawecki TJ (1998) Red queen meets Santa Rosalia: Arms races and the evolution of host specialization in organisms with parasitic lifestyles. Am Nat 152: 635–651. [DOI] [PubMed] [Google Scholar]
- 8.Begon M, Townsend CR, Harper JL (2006) Ecology: From individuals to ecosystems. Oxford: Blackwell Publishing. 752 p.
- 9.Cope ED (1903) The primary factors of organic evolution. Chicago: Open Court Publishing Company. 547 p.
- 10. Johnson KP, Adams RJ, Page RDM, Clayton DH (2003) When do parasites fail to speciate in response to host speciation? Syst Biol 52: 37–47. [DOI] [PubMed] [Google Scholar]
- 11. Hoberg EP, Brooks DR (2008) A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host-parasite systems. J Biogeogr 35: 1533–1550. [Google Scholar]
- 12. Janz N, Nylin S (1998) Butterflies and plants: A phylogenetic study. Evolution 52: 486–502. [DOI] [PubMed] [Google Scholar]
- 13. Crespi BJ, Sandoval CP (2000) Phylogenetic evidence for the evolution of ecological specialization in Timema walking-sticks. J Evol Biol 13: 249–262. [Google Scholar]
- 14. Scheffer SJ, Wiegmann BM (2000) Molecular Phylogenetics of the Holly Leafminers (Diptera: Agromyzidae: Phytomyza): Species Limits, Speciation, and Dietary Specialization. Mol Phylogenet Evol 17: 244–255. [DOI] [PubMed] [Google Scholar]
- 15. Janz N, Nyblom K, Nylin S (2001) Evolutionary dynamics of host-plant specialization: A case study of the tribe Nymphalini. Evolution 55: 783–796. [DOI] [PubMed] [Google Scholar]
- 16. Ronquist F, Liljeblad J (2001) Evolution of the gall wasp-host plant association. Evolution 55: 2503–2522. [DOI] [PubMed] [Google Scholar]
- 17. Lopez-Vaamonde C, Charles H, Godfray HCJ, Cook JM (2003) Evolutionary dynamics of host-plant use in a genus of leaf-mining moths. Evolution 57: 1804–1821. [DOI] [PubMed] [Google Scholar]
- 18. Favret C, Voegtlin DJ (2004) Speciation by host-switching in pinyon Cinara (Insecta: Hemiptera: Aphididae). Mol Phylogenet Evol 32: 139–151. [DOI] [PubMed] [Google Scholar]
- 19. Poulin R, Krasnov BR, Shenbrot GI, Mouillot D, Khokhlova IS (2006) Evolution of host specificity in fleas: Is it directional and irreversible? Int J Parasitol 36: 185–191. [DOI] [PubMed] [Google Scholar]
- 20. Desdevises Y, Morand S, Legendre P (2002) Evolution and determinants of host specificity in the genus Lamellodiscus (Monogenea). Biol J Linn Soc Lond 77: 431–443. [Google Scholar]
- 21. Janzen DH (1985) On ecological fitting. Oikos 45: 308–310. [Google Scholar]
- 22. Agosta SJ, Klemens JA (2008) Ecological fitting by phenotypically flexible genotypes: implications for species associations, community assembly and evolution. Ecol Lett 11: 1123–1134. [DOI] [PubMed] [Google Scholar]
- 23. Robinson BW, Wilson DS (1998) Optimal foraging, specialization, and a solution to Liem's paradox. Am Nat 151: 223–235. [DOI] [PubMed] [Google Scholar]
- 24.Michener CD (2007) The bees of the world: Baltimore: Johns Hopkins University Press. 953 p.
- 25. Williams NM (2003) Use of novel pollen species by specialist and generalist solitary bees (Hymenoptera: Megachilidae). Oecologia 134: 228–237. [DOI] [PubMed] [Google Scholar]
- 26. Praz CJ, Mueller A, Dorn S (2008) Specialized bees fail to develop on non-host pollen: Do plants chemically protect their pollen? Ecology 89: 795–804. [DOI] [PubMed] [Google Scholar]
- 27. Sedivy C, Muller A, Dorn S (2011) Closely related pollen generalist bees differ in their ability to develop on the same pollen diet: evidence for physiological adaptations to digest pollen. Funct Ecol 25: 718–725. [Google Scholar]
- 28. Bernays EA, Wcislo WT (1994) Sensory capabilities, information processing, and resource specialization. Q Rev Biol 69: 187–204. [Google Scholar]
- 29. Sedivy C, Praz CJ, Muller A, Widmer A, Dorn S (2008) Patterns of host-plant choice in bees of the genus Chelostoma: the constraint hypothesis of host-range evolution in bees. Evolution 62: 2487–2507. [DOI] [PubMed] [Google Scholar]
- 30. Muller A (1996) Host-plant specialization in western Palearctic anthidine bees (Hymenoptera: Apoidea: Megachilidae). Ecol Monogr 66: 235–257. [Google Scholar]
- 31. Michez D, Patiny S, Rasmont P, Timmermann K, Vereecken NJ (2008) Phylogeny and host-plant evolution in Melittidae s.l. (Hymenoptera: Apoidea). Apidologie 39: 146–162. [Google Scholar]
- 32. Chittka L, Thomson JD, Waser NM (1999) Flower constancy, insect psychology, and plant evolution. Naturwissenschaften 86: 361–377. [Google Scholar]
- 33. Michener CD (1956) The ethology of Andrena erythronii with comparative data on other species (Hymenoptera, Andrenidae). Sci Pap Univ Kansas Nat Hist Mus 37: 645–684. [Google Scholar]
- 34. Westrich P (2008) Flexibles Pollensammelverhalten der ansonsten streng oligolektischen Seidenbiene Colletes hederae Schmidt & Westrich (Hymenoptera: Apidae). Eucera 1: 17–32. [Google Scholar]
- 35. Straka J, Bogusch P (2007) Phylogeny of the bees of the family Apidae based on larval characters with focus on the origin of cleptoparasitism (Hymenoptera: Apiformes). Syst Entomol 32: 700–711. [Google Scholar]
- 36. Cardinal S, Straka J, Danforth BN (2010) Comprehensive phylogeny of apid bees reveals the evolutionary origins and antiquity of cleptoparasitism. Proc Natl Acad Sci U S A107: 16207–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogusch P (2003) Biology of selected kleptoparasitic bee species. Prague: Charles University in Prague. 96 p.
- 38. Bogusch P, Kratochvil L, Straka J (2006) Generalist cuckoo bees (Hymenoptera: Apoidea: Sphecodes) are species-specialist at the individual level. Behav Ecol Sociobiol 60: 422–429. [Google Scholar]
- 39. Bogusch P, Straka J (2012) Review and identification of the cuckoo bees of central Europe (Hymenoptera: Halictidae: Sphecodes). Zootaxa 3311: 1–41. [Google Scholar]
- 40. Michener CD (1978) The parasitic groups of Halictidae (Hymenoptera, Apoidea). Sci Pap Univ Kansas Nat Hist Mus 51: 291–339. [Google Scholar]
- 41.Griekspoor A, Groothuis T (2006) 4Peaks. Available: mekentosj.com.
- 42. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 43. Villesen P (2007) FaBox: an online toolbox for FASTA sequences. Mol Ecol Notes 7: 965–968. [Google Scholar]
- 44. Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19: 716–723. [Google Scholar]
- 45. Sugiura N (1978) Further analysts of the data by akaike's information criterion and the finite corrections. Commun Stat Theory Methods 7: 13–26. [Google Scholar]
- 46. Hurvich CM, Tsai CL (1989) Regression and time series model selection in small samples. Biometrika 76: 297–307. [Google Scholar]
- 47. Posada D (2008) jModelTest: Phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 48. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 49. Kass RE, Raftery AE (1995) Bayes Factors. J Am Stat Assoc 90: 773–795. [Google Scholar]
- 50. Brandley MC, Schmitz A, Reeder TW (2005) Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol 54: 373–390. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Skjæveland Å, Orr RJ, Enger P, Ruden T, et al.. (2009) AIR: A batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics 10. [DOI] [PMC free article] [PubMed]
- 52.Rambaut A, Drummond A (2007) Tracer v1.4. Available: http://beast.bio.ed.ac.uk/Tracer. Accessed 2012 Dec 12.
- 53.Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: The University of Texas at Austin. 115 p.
- 54.Swofford DL (1998) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, Massachusetts: Sinauer Associates.
- 55.Blüthgen P (1934) Die Wirte der Paläarktischen Sphecodes-Arten. Zeitschrift Wissenschaftlicher Insekten-Biologie 27: 33–42, 61–66.
- 56. Danforth BN (1989) Nesting behavior of four species of Perdita (Hymenoptera: Andrenidae). J Kansas Entomol Soc 62: 59–79. [Google Scholar]
- 57. Eickwort GC (1977) Aspects of the nesting biology and descriptions of immature stages of Perdita octomaculata and P. halictoides (Hymenoptera: Andrenidae). J Kansas Entomol Soc 50: 577–599. [Google Scholar]
- 58. Gibbs J, Brady SG, Kanda K, Danforth BN (2012) Phylogeny of halictine bees supports a shared origin of eusociality for Halictus and Lasioglossum (Apoidea: Anthophila: Halictidae). Mol Phyl Evol 65: 926–939. [DOI] [PubMed] [Google Scholar]
- 59. Rohde K (1980) Host specificity indices of parasites and their application. Experientia 36: 1369–1371. [Google Scholar]
- 60. Poulin R, Mouillot D (2003) Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology 126: 473–480. [DOI] [PubMed] [Google Scholar]
- 61.Poulin R (2007) Evolutionary Ecology of Parasites. Princeton: Princeton University Press. 332 p.
- 62. Pagel M, Meade A, Barker D (2004) Bayesian estimation of ancestral character states on phylogenies. Syst Biol 53: 673–684 doi:10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 63.Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75. Available: http://mesquiteproject.org. Accessed 2012 Dec 12.
- 64. Pagel M, Meade A (2006) Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat 167: 808–825 doi:10.2307/3844739. [DOI] [PubMed] [Google Scholar]
- 65. Engel MS (2006) A new genus of cleptoparasitic bees from the West Indies (Hymenoptera: Halictidae). Acta Zool Crac 49B: 1–8. [Google Scholar]
- 66.Rasmussen C, Michener CD (2011) Callosphecodes, a little-known bee (Hymenoptera, Halictidae, Sphecodes). Zookeys: 61–68. [DOI] [PMC free article] [PubMed]
- 67. Robertson C (1903) Synopsis of Sphecodinae. Entomol News 14: 103–107. [Google Scholar]
- 68. Emery C (1909) Über den Ursprung der dulotischen, parasitischen und myrmekophilen Ameisen Biol Zent Bl. 29: 352–362. [Google Scholar]
- 69. Carpenter JM, Strassmann JE, Turillazzi S, Hughes CR, Solis CR, et al. (1993) Phylogenetic Relationships Among Paper Wasp Social Parasites and Their Hosts (Hymenoptera: Vespidae; Polistinae). Cladistics 9: 129–146. [DOI] [PubMed] [Google Scholar]
- 70. Gibbs J, Albert J, Packer L (2012) Dual origins of social parasitism in North American Dialictus (Hymenoptera: Halictidae) confirmed using a phylogenetic approach. Cladistics 28: 195–207 doi:10.1111/j.1096-0031.2011.00373.x. [DOI] [PubMed] [Google Scholar]
- 71. Almeida EAB, Pie MR, Brady SG, Danforth BN (2012) Biogeography and diversification of colletid bees (Hymenoptera: Colletidae): emerging patterns from the southern end of the world. J Biogeogr 39: 526–544. [Google Scholar]
- 72.Westrich P (1989) Die Wildbienen Baden-Württembergs. Stuttgart: E. Ulmer.972 p.
- 73.Rozen JG (1965) The biology and immature stages of Melitturga clavicornis (Latreille) and of Sphecodes albilabris (Kirby) and the recognition of the Oxaeidae at the family level (Hymenoptera, Apoidea). Am Mus Novit 2224.
- 74. Agosta SJ, Janz N, Brooks DR (2010) How specialists can be generalists: resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia (Curitiba) 27: 151–162. [Google Scholar]
- 75. Rezkova K, Žáková M, Žáková Z, Straka J (2012) Analysis of nesting behavior based on daily observation of Andrena vaga (Hymenoptera: Andrenidae). J Insect Behav 25: 24–47. [Google Scholar]
- 76. Sick M, Ayasse M, Tengö J, Engels W, Lübke G, et al. (1994) Host-parasite relationships in six species of Sphecodes bees and their halictid hosts: Nest intrusion, intranidal behavior, and Dufour's gland volatiles (Hymenoptera: Halictidae). J Insect Behav 7: 101–117. [Google Scholar]
- 77. Packer L, Dzinas A, Strickler K, Scott V (1995) Genetic differentiation between two host “races” and two species of cleptoparasitic bees and between their two hosts. Biochem Genet 33: 97–109. [DOI] [PubMed] [Google Scholar]
- 78. Danforth BN, Sipes S, Fang J, Brady SG (2006) The history of early bee diversification based on five genes plus morphology. Proc Natl Acad Sci U S A103: 15118–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Danforth BN (1999) Phylogeny of the bee genus Lasioglossum (Hymenoptera: Halictidae) based on mitochondrial COI sequence data. Syst Entomol 24: 377–393. [Google Scholar]
- 80. Danforth BN, Sauquet H, Packer L (1999) Phylogeny of the bee genus Halictus (Hymenoptera: Halictidae) based on parsimony and likelihood analyses of nuclear EF-1 alpha sequence data. Mol Phylogenet Evol 13: 605–618. [DOI] [PubMed] [Google Scholar]
- 81. Danforth BN, Brady SG, Sipes SD, Pearson A (2004) Single-copy nuclear genes recover Cretaceous-age divergences in bees. Syst Biol 53: 309–326. [DOI] [PubMed] [Google Scholar]
- 82. Danforth BN, Eardley C, Packer L, Walker K, Pauly A, et al. (2008) Phylogeny of Halictidae with an emphasis on endemic African Halictinae. Apidologie (Celle) 39: 86–U42. [Google Scholar]
- 83. Danforth BN (2002) Evolution of sociality in a primitively eusocial lineage of bees. Proc Natl Acad Sci USA 99: 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Danforth BN, Fang J, Sipes S (2006) Analysis of family-level relationships in bees (Hymenoptera: Apiformes) using 28S and two previously unexplored nuclear genes: CAD and RNA polymerase II. Mol Phylogenet Evol 39: 358–372. [DOI] [PubMed] [Google Scholar]
- 85. Sheffield CS, Hebert PDN, Kevan PG, Packer L (2009) DNA barcoding a regional bee (Hymenoptera: Apoidea) fauna and its potential for ecological studies. Mol Ecol Resour 9: 196–207. [DOI] [PubMed] [Google Scholar]
- 86. Soucy SL, Danforth BN (2002) Phylogeography of the socially polymorphic sweat bee Halictus rubicundus (Hymenoptera: Halictidae). Evolution 56: 330–341. [DOI] [PubMed] [Google Scholar]
- 87. Danforth BN, Conway L, Ji SQ (2003) Phylogeny of eusocial Lasioglossum reveals multiple losses of eusociality within a primitively eusocial clade of bees (Hymenoptera: Halictidae). Syst Biol 52: 23–36. [DOI] [PubMed] [Google Scholar]
- 88. Danforth BN, Ji SQ (2001) Australian Lasioglossum plus Homalictus form a monophyletic group: Resolving the “Australian enigma”. Syst Biol 50: 268–283. [PubMed] [Google Scholar]
- 89. Pedersen BV (1996) A phylogenetic analysis of cuckoo bumblebees (Psithyrus, Lepeletier) and bumblebees (Bombus, Latreille) inferred from sequences of the mitochondrial gene cytochrome oxidase. Mol Phylogenet Evol 5: 289–297. [DOI] [PubMed] [Google Scholar]
- 90. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87: 651–701. [Google Scholar]
- 91. Schulmeister S (2003) Simultaneous analysis of basal Hymenoptera (Insecta): introducing robust-choice sensitivity analysis. Biol J Linn Soc Lond 79: 245–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree resulting from Bayesian analysis of a complete dataset (Align2). Posterior probabilities are in front of the slash; bootstrap values of the maximum likelihood analysis are behind the slash. Posterior probabilities lower than 0.5 and bootstrap values lower than 50 are replaced by an asterisk (*).
(TIF)
Character state reconstruction of host specificity in cuckoo bees of the genus Sphecodes . The larger pie charts show posterior probabilities obtained by Bayesian analysis using Distribution II (main analysis). The smaller pie charts show the results (posterior probabilities in Bayesian analyses or possible character states in maximum parsimony analyses) of other analyses in which interpretations differed from those of the main analysis. B1, Bayesian analysis using Distribution I; MP1, maximum parsimony analysis using Distribution I; B3, Bayesian analysis using Distribution III; and MP3, maximum parsimony analysis using Distribution III.
(TIF)
Complete results of character state reconstruction of host specifity. DI: Distribution of specialist and generalist I, DIII: Distribution of specialist and generalist III, P(G): posterior probability for generalism as ancestral state, P(S): posterior probability for specialism as ancestral state, BF: Bayes Factor (support of more probable state). Values of BF lower than 2 are replaced by asterisk (*).
(DOC)
Complete results of character state reconstruction of ancestral hosts. P (LS): posterior probability for Lasioglossum s. s. as ancestral host, P (LO): posterior probability for Lasioglossum other as ancestral host, P (H): posterior probability for Halictus subgenus Halictus as ancestral host, P (S): posterior probability for Halictus subgenus Seladonia as ancestral host, P (A) posterior probability for Andrena as ancestral host, P (C): posterior probability for Colletes as ancestral host, P (P): posterior probability for Perdita as ancestral host, P (M): posterior probability for Melitturga as ancestral host. The values higher than 0.7 are in bold.
(DOC)