Abstract
Plant ATP-binding cassette (ABC) transporters consist of largest family members among many other membrane transporters and have been implicated in various functions such as detoxification, disease resistance and transport of diverse substrates. Of the ABC-B/multi-drug resistance/P-glycoprotein (ABCB/MDR/PGP) subfamily, at least five members have been reported to mediate cellular transport of auxin or auxin derivatives. Although single mutant phenotypes of these genes are milder than PIN-FORMED (PIN) mutants, those ABCBs significantly contribute for the directional auxin movement in the tissue-level auxin-transporting assay. Uniformly localized ABCB proteins in the plasma membrane (PM) are generaly found in different plant species and stably retained regardless of internal and external signals. This implies that these ABCB proteins may play as basal auxin transporters.
Keywords: ABCB, PGP, auxin transporter, non-polar localization, sterol, PIN, Arabidopsis
Auxin is a key hormone regulating plant growth and development. Auxin gradients formed by directional auxin transport are critical for the biological functions of auxin. Directional auxin transport and auxin gradient formation across tissues are accomplished by cooperation of different auxin transporters. So far, three types of auxin transporters have been reported; AUXIN RESISTANT 1/LIKE AUX1s (AUX1/LAXs) as auxin influx carriers, PINs and ABCB/MDR/PGPs as auxin efflux carriers.1,2
The plant ABCB subfamily includes 21 members in three clusters. These transporter proteins consist of two similar halves connected by a linker in which each half contains a transmembrane domain (TMD) and a nucleotide-binding domain (NBD).3 Among the ABCB subfamily, ABCB1, ABCB4 and ABCB19 have been well characterized as auxin transporters.4 However, recent studies showed that other ABCBs like ABCB14 and ABCB15 also are associated with polar auxin transport.5 All PINs are involved in auxin transport and share functional similarity within the subgroup. However, only a subset of ABCBs has been reported to transport auxin, and those auxin-transporting ABCBs (AT-ABCBs) do not belong to the same phylogenetic clade; namely, ABCB1, ABCB14 and ABCB19 to clade I, ABCB4 to clade II, and ABCB15 to clade III.
Single mutants of AT-ABCBs generally show milder phenotypes than those of PINs. The mdr1 mutants showed epinasty of cotyledons and first true leaves and waviness in hypocotyl and roots.6 Defects of ABCB4 grew longer root hairs than wild type,7 and the abcb14 mutants exhibited slightly altered vascular development in the florescence stem.5 The abcb1 abcb19 double mutant showed a dwarfism.6 In the auxin transport assay using Arabidopsis root hairs,8 root-hair-specifically overexpressed AT-ABCBs mildly reduced root hair growth, whereas overexpression of PINs, except PIN5, showed great effects on root hair inhibition.7,9 Based on the fact that auxin positively regulates root hair growth, ABCB1, 4, and19 seem to have lower auxin-exporting capability than PINs.
Despite their relatively weak auxin-exporting activity in root hair cells, single mutations of AT-ABCBs considerably affected polar auxin transport. ABCB19 localizes to the vascular tissues of the hypocotyl and to the stele of the root.1 ABCB19 localization in the apical tissues suggests its role for auxin loading into the basipetal stream in both shoot and root tips.1 ABCB14 and ABCB15 also localize mainly to the vascular tissues of the inflorescence stem and affect polar auxin transport in those tissues.5 Conversely, ABCB1 and ABCB4 facilitate shootward auxin transport. ABCB1 expresses in the stele tissues of the root apical region (between the lateral root cap and the elongation zone) and in root cortical and epidermis cells and is involved in loading and transport of auxin into the shootward stream from the root apex.10 ABCB4, being expressed in the root epidermis, also mediates shootward auxin transport.11
Mutants of abcb19 and abcb1 abcb19 reduced the rootward auxin transport more greatly than did pin1.12 This is consistent with the observation that mis-localized ABCB proteins in the twd1 (TWISTED DWARF1) mutant displayed only 14% of wild-type polar auxin transport activity.13 Considering the non-polar property of ABCB19, this result is intriguing. The recent report suggests a model how ABCB19 affects PIN-mediated directional auxin transport. ABCB19 localizes to the detergent-resistant membrane (DRM) of the PM where sitosterol and glucosylceramide are abundant.14 ABCB19 does not alter the polar localization of PIN1 but stabilizes PIN1 in the DRM.14 Titapiwatanakun et al. suggested that ABCB19 defines the discrete membrane structure such as DRM and provides a platform for stable PIN1 localization so as to affect the PIN trafficking and function.14 In the abcb19 mutant, the endocytic tracer FM4–64 was more diffusive than in wild type, and brefeldin A (BFA) bodies including PIN1 did not fully disappeared even after BFA washout.14 These observations imply that ABCB19, in addition to its direct auxin transport activity, might have other function in maintaining the membrane structure for PIN1. Destabilization of PIN1 by the loss of ABCB19 showed overall less polar auxin transport activity than did the pin1 mutant.14 PIN localization is sensitively affected by sterol composition.15,16 A recent study has suggested that clustering of PIN proteins in the PM is important to maintain their polarity in addition to super-polar exocytosis and endocytosis.17 PIN2 is associated with the cluster at the lateral side of PM to prevent lateral diffusion.17 Filipin, the sterol-binding agent, reduced PIN2 clustering and subsequently increased lateral diffusion.17 This result implies that sterols contribute for maintaining PINs polarity and their function as well. DRM is the sterol-rich PM compartment which can be disturbed by filipin and cyclodextrin. Other ABCB proteins as well as ABCB19 might play a role for the association of PINs into the cluster and/or are possibly involved in intracellular distribution and transport of sterols into the PM, which will ultimately contribute to confine the discrete microdomains. It has been demonstrated in yeast that sterol transport from ER to PM by ABC proteins regulates sterol compositions in the PM.18,19 Interestingly, ABCB14 has been known as a malate importer modulating stomatal movement in guard cells.20 Furthermore, ABCB4 has recently been suggested to be a target of herbicidal activity by 2,4-D, a non-competitive inhibitor of IAA transport.21 These results open the possibility that ABCB proteins have multiple molecular functions.
ABCB proteins localize to the PM largely in a non-polar manner. While PINs trafficking pathways have been intensively studied due to their polar localization dynamicity in response to developmental and environmental cues, ABCBs trafficking pathways have been relatively less investigated. ABCB1, ABCB4 and ABCB19 require TWD1 for their targeting to the PM.22 However, once they have targeted to the PM, AT-ABCBs stably anchor in the PM and barely change their localization in response to internal and external cues. AT-ABCBs’ stability in the PM and differential regulatory mechanisms are supported by their resistance to BFA and non-recycling nature between PM and early endosomes.13,23 However, ABCB4 has recently been shown to share the degradation pathway with PINs but with much slower endocytic rate.23 Uniformly localized and non-dynamic AT-ABCB proteins may serve as basal auxin transporters for the regulation of cellular auxin homeostasis and fine tuning of auxin distribution by working together with PINs. Highly conserved structures of AT- ABCBs throughout the plant and their weak auxin-exporting activities support this hypothesis.1
ABCB1, ABCB4 and ABCB19, albeit with the structural similarity with human MDRs, have been known to facilitate auxin transport. ABCB14 and ABCB15 also have been additionally known to mediate auxin transport. These imply that more ABCB proteins could be involved in auxin transport. On the other hand, functional activities of ABCB4, ABCB14 and ABCB19 other than auxin transport have also been demonstrated. This raises a question whether AT-ABCB proteins had originally evolved as auxin transporters or they had gained the function later. To more accurately understand the contribution of AT-ABCB proteins in auxin transport, other ABCB family proteins also need to be functionally characterized. (Author, please cite Figs. 1 and 2 appropriately in the main text)
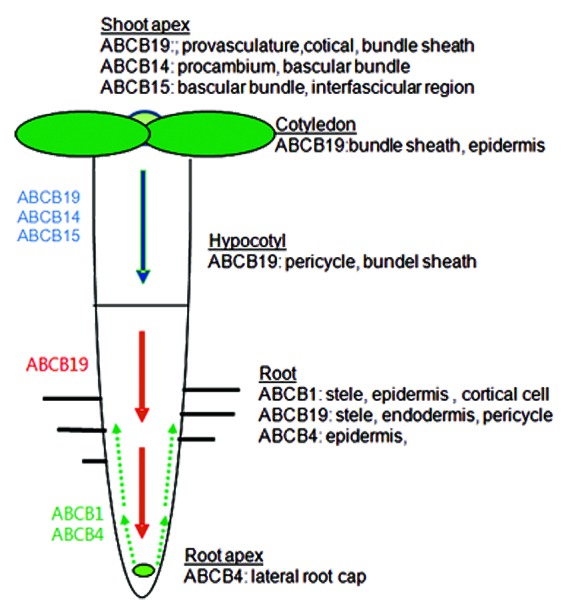
Figure 1. Expression and contribution of ABCB proteins for the polar auxin transport.
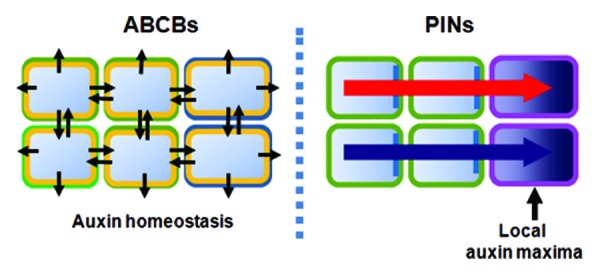
Figure 2. Distinctive functions between ABCBs and PINs in auxin transport. ABCBs, localizing stably and symmetrically in the PM, function as basal auxin transporters and regulate cellular auxin homeostasis. In contrast, PINs, asymmetrically localized and with distribution dynamicity in response to developmental and environmental cues, mediate directional auxin transport to raise formative local auxin gradients.
Acknowledgments
This work was supported by grants from the Mid-career Researcher Program (2011–0017242, NRF, MEST) and the Next-Generation BioGreen 21 programs (TAGC PJ00820701 and SSAC PJ00814102) of the Rural Development Administration.
Glossary
Abbreviations:
- ABCB
ATP-binding cassette class B
- AUX1
AUXIN RESISTANT
- BFA
Brefeldin A1
- DRM
detergent-resistant membrane
- IAA
indole-3-acetic acid
- LAX
LIKE AUX1
- MDR
multi-drug resistance
- NBD
nucleotide binding domain
- PGP
P-glycoprotein
- PIN
PIN-FORMED
- TMD
transmembrnae domain
- TWD1
TWISTED DWARF1
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22990
References
- 1.Zazímalová E, Murphy AS, Yang H, Hoyerová K, Hosek P. Auxin transporters--why so many? Cold Spring Harb Perspect Biol. 2010;2:a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–88. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 3.Geisler M, Murphy AS. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 2006;580:1094–102. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- 4.Titapiwatanakun B, Murphy AS. Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot. 2009;60:1093–107. doi: 10.1093/jxb/ern240. [DOI] [PubMed] [Google Scholar]
- 5.Kaneda M, Schuetz M, Lin BS, Chanis C, Hamberger B, Western TL, et al. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot. 2011;62:2063–77. doi: 10.1093/jxb/erq416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13:2441–54. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho M, Lee SH, Cho H-T. P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell. 2007;19:3930–43. doi: 10.1105/tpc.107.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SH, Cho H-T. Auxin and the Root Hair Morphogenesis. In Root Hairs. 2009; AM Emons and T Ketelaar (ed.) Springer Publishers, Berlin. [Google Scholar]
- 9.Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho H-T. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 2010;153:1046–61. doi: 10.1104/pp.110.156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–94. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 11.Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell. 2007;19:1838–50. doi: 10.1105/tpc.107.051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19:131–47. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell. 2003;14:4238–49. doi: 10.1091/mbc.E02-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, et al. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- 15.Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B. Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell. 2003;15:612–25. doi: 10.1105/tpc.008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, et al. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol. 2008;10:237–44. doi: 10.1038/ncb1686. [DOI] [PubMed] [Google Scholar]
- 17.Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol. 2011;7:540. doi: 10.1038/msb.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Prinz WA. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J Biol Chem. 2004;279:45226–34. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 19.Cabrito TR, Teixeira MC, Singh A, Prasad R, Sá-Correia I. The yeast ABC transporter Pdr18 (ORF YNR070w) controls plasma membrane sterol composition, playing a role in multidrug resistance. Biochem J. 2011;440:195–202. doi: 10.1042/BJ20110876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M, Choi Y, Burla B, Kim YY, Jeon B, Maeshima M, et al. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat Cell Biol. 2008;10:1217–23. doi: 10.1038/ncb1782. [DOI] [PubMed] [Google Scholar]
- 21.Kubeš M, Yang H, Richter GL, Cheng Y, Młodzińska E, Wang X, et al. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012;69:640–54. doi: 10.1111/j.1365-313X.2011.04818.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Otegui MS, Spalding EP. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell. 2010;22:3295–304. doi: 10.1105/tpc.110.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho M, Lee ZW, Cho HT. ATP-binding cassette B4, an auxin-efflux transporter, stably associates with the plasma membrane and shows distinctive intracellular trafficking from that of PIN-FORMED proteins. Plant Physiol. 2012;159:642–54. doi: 10.1104/pp.112.196139. [DOI] [PMC free article] [PubMed] [Google Scholar]


