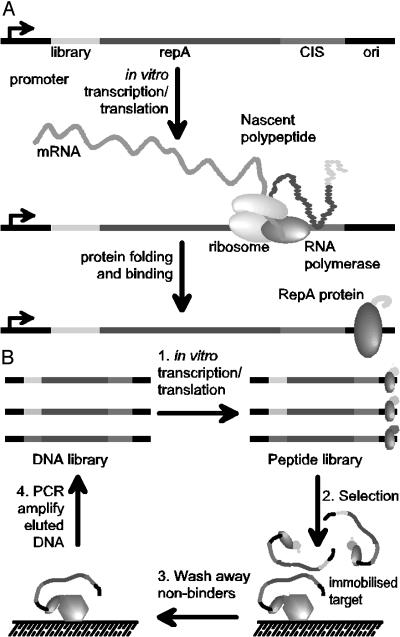
Fig. 1.
The principle of the CIS display technology. (A) Template DNA encoding an N-terminal library peptide is ligated to the RepA gene. In vitro transcription is initiated at the promoter and pauses when the RNA polymerase reaches the CIS element. Concurrent translation produces the RepA protein, which transiently interacts with the CIS element, thereby forcing its subsequent binding to the adjacent ori sequence. This process establishes a faithful linkage between a template DNA and the expressed polypeptide that it encodes. (B) CIS display selections begin with the construction of a peptide-encoding DNA library followed by in vitro transcription/translation to form a pool of protein–DNA complexes (step 1). The library pool is incubated with an immobilized target (step 2), nonbinding peptides are washed away (step 3), and the retained DNA that encodes the target-binding peptides is eluted and amplified by PCR (step 4), to form a DNA library ready for the next round of selection. After three to five rounds of selection, recovered DNA is cloned into an appropriate expression vector for the identification of individual target-binding peptide sequences.