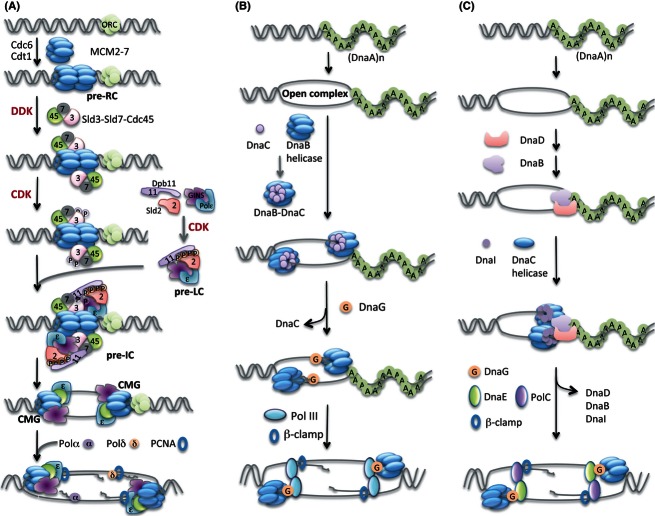
Figure 1.
Origin loading and activation of replicative helicases. (A) In eukaryotes, the first step is loading of Mcm2–7 helicase onto the replication origin region that depends on origin recognition heterohexamer ORC and two loading proteins Cdc6 and Cdt1. In budding yeast S. cerevisiae, the subsequent activation of helicase requires DDK and CDK, recruiting other replication initiation proteins including a Sld3-Sld7-Cdc45 complex and then a preloading complex (pre-LC) composed of Sld2, Dpb11, GINS tetramer and polymerase DNA Pol ε onto the head-to-head double MCM hexamers to form a huge pre-initiation complex (pre-IC) at origin regions, immediately followed by remodeling that leads to the activation of CMG helicase. This process probably switches MCM from encircling dsDNA to ssDNA. Next, the active CMG works together with DNA polymerases, Pol α, Pol δ and Pol ε to duplicate DNA. (B) In Gram-negative E. coli, the initiator DnaA recognizes the bacterial replication origin oriC and melts a specific origin sequence to form an open complex in the presence of ATP. Then, DnaB–DnaC complex is loaded on each separated single-stranded DNA. The binding of primase DnaG with DnaB helicase and the primer synthesis induce removal of DnaC from DnaB. The DnaC dissociation requires ATPase activity. Subsequent loading of DNA replicative polymerase Pol III and β-clamp triggers replication elongation. (C) In Gram-positive B. subtilis, DnaA binds and melts oriC sequence, followed by loading of DnaD and DnaB orderly onto the origin region. DnaC helicase and DnaI assemble to the origin subsequently in the presence of ATP. The remodeling including removal of DnaD, DnaB and DnaI and the assembly of primase DnaG, two DNA polymerases, DnaE and PolC, and β-clamp activates helicase and replication fork movement.