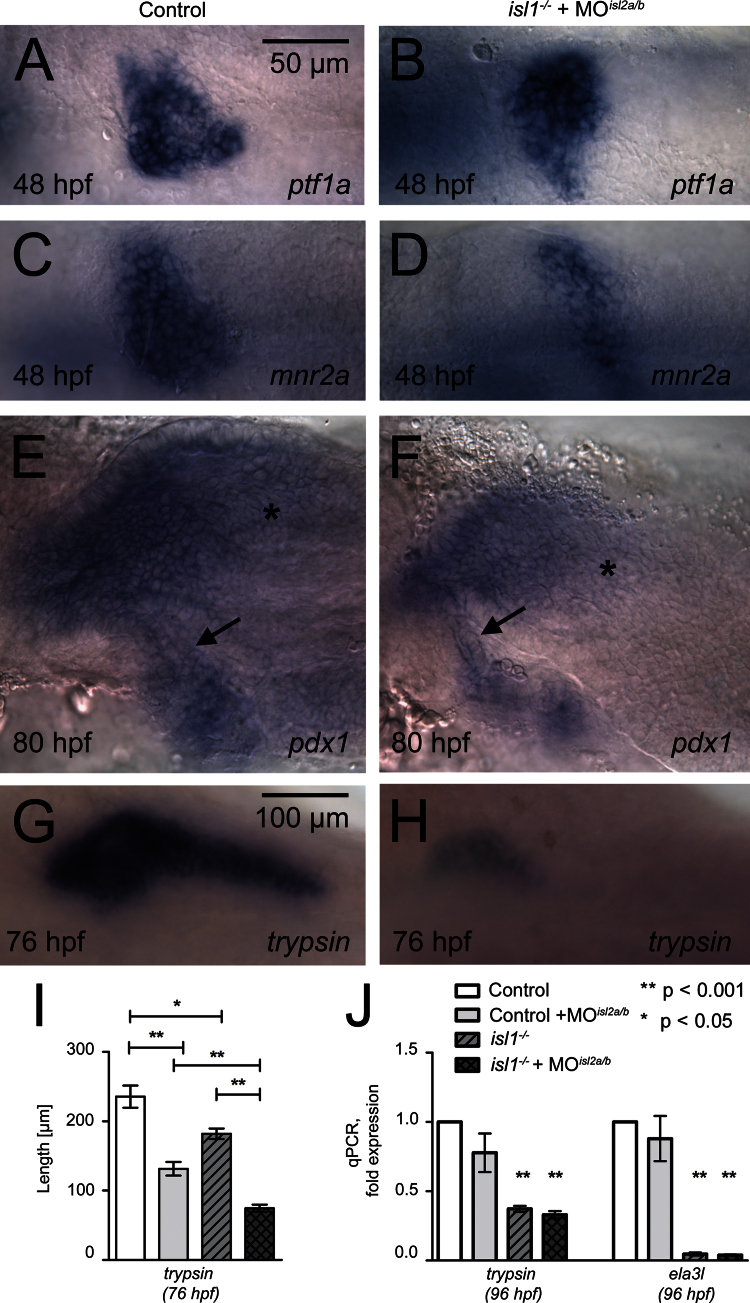
Fig. 5.
Overlapping activities of three isl-genes in exocrine tissue formation. (A, B) Pancreatic ptf1/p48 mRNA expression is similar in control (A) and isl1−/−+MOisl2a/2b embryos (B) at 48 hpf. (C, D) Differently, pancreatic mnr2a mRNA expression is reduced in isl1−/−+MOisl2a/2b embryos as compared to control embryos. (E, G) pdx1 mRNA expression at 80 hpf reveals a similar morphology of the extrapancreatic duct (arrow) in control and isl1−/−+MOisl2a/2b embryos. Note the reduced gut specific pdx1 signals (asterisk) in the isl-depleted embryos. (G, H) trypsin mRNA expressing in exocrine tissue of a control embryo (G) and in an isl1−/−+MOisl2a/2b embryos (G) at 76 hpf. (I) Quantitative analyses on the length of trypsin mRNA expression domain in control and triple isl gene deficient embryos at 76 hpf. Embryos are shown from ventral with anterior to the left (A–F) or dorsal with anterior to the left (G, H). P-values for significant changes are indicated. (J) qPCR analysis for mRNA expression levels of trypsin and ela3l in 96 hpf old whole embryos, normalized to EF1α. Coloring of the bars corresponds to the same genotypes presented in (I). Bars show mean+SEM.