Figure 3.
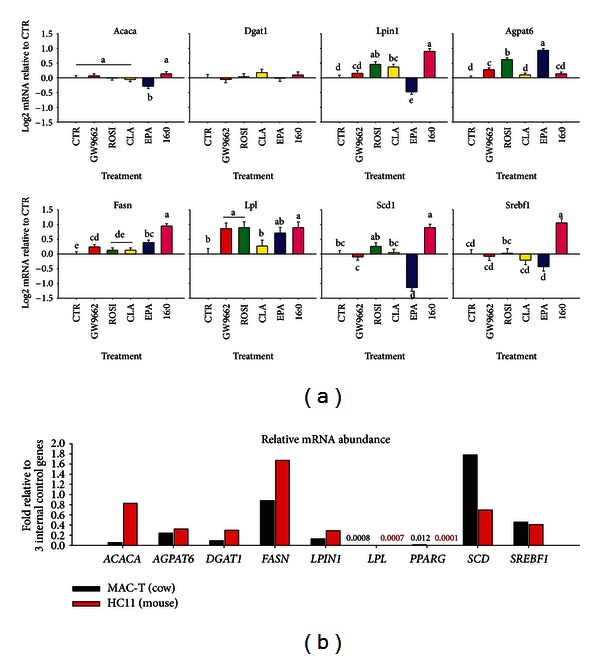
Effect of PPARγ activation on genes coding for proteins involved in milk fat synthesis in mouse mammary epithelial cells HC11. The experiment was performed with the purpose to test the effects of 50 μM of the PPARγ activator rosiglitazone, the PPARγ inhibitor GW9662, or 100 μM of several long-chain fatty acids (trans-10,cis 12-conjugated linoleic acid (CLA), eicosapentaenoic acid (EPA), or palmitate (16:0)) for 12 hours in HC11 cells and compare the data with results using the same experimental design (except the GW9662 treatment) in MAC-T cells [26]. All the procedures with few modifications were as previously described [26]. The RNA was extracted and qPCR performed for several genes known to be involved in milk fat synthesis and significantly upregulated by rosiglitazone in MAC-T cells and the same 3 internal control genes used [26]. In (a), the effect of treatments on HC11 cell is reported. For that experiment, the qPCR data were calculated as fold change relative to control and log2 transformed prior statistical analysis using Proc GLM of SAS with treatment as main effect and replicate as random. Dissimilar letters denote significant differences between treatments (P < 0.05). In (b), a comparison in mRNA abundance between measured genes in the control group of HC11 and MAC-T cells is presented. The relative mRNA abundance was calculated as previously described [26] but as fold difference relative to the geometric mean of the median Ct values of the 3 internal control genes instead as % relative abundance. The same analysis was performed for the MAC-T cells using data previously published [26]. The PPARG was detectable only for few samples in HC11 cells and LPL was barely detectable in both HC11 and MAC-T cells.
