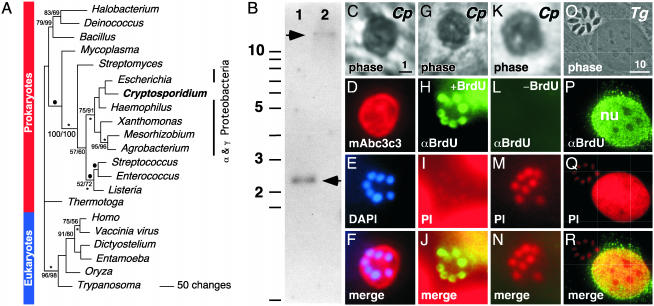
Fig. 3.
C. parvum expresses a TK of eubacterial origin. (A) Phylogenetic analysis of TK. The tree shown is one of four most parsimonious trees obtained. Solid bullets indicate branches not found on the other most parsimonious trees. Numbers above the branches (space permitting) indicate the bootstrap values for parsimony and neighbor-joining analyses. Only percentages >50% are shown. Asterisks below branches indicate nodes with ≥50% frequency support from maximum likelihood puzzle analysis. GenBank accession numbers, multiple sequence alignment, and analysis details are available in Materials and Methods and supporting information. Many organisms lack a TK gene including δ- and ε-proteobacteria. (B) Genomic Southern blot of C. parvum type 2 DNA probed with TK coding sequence. Lane 1, EcoRI; lane 2, BamHI. Sizes are as indicated (expected sizes based on genome sequence data are 2.4 and >11.5 kb, respectively). (C) C. parvum type 1 meronts were identified in infected tissue cultures based on their reactivity with the C. parvum-specific mAb c3c3 (D) and the presence of six to eight nuclei (E, I, and M). These nuclei could be detected after BrdUrd labeling with an antibody specific for incorporated BrdUrd (H). No antibody labeling was observed without prior BrdUrd labeling (L) or in T. gondii (P), which lacks a TK gene. DAPI, 6′-diamidino-2-phenylinidole; PI, propidium iodide; Cp, C. parvum; Tg, T. gondii; nu, host cell nucleus. [Scale bar represents 1 (C-N) or 10 μm (O-R), respectively.]