Abstract
Targeting noncatalytic cysteine residues with irreversible acrylamide-based inhibitors is a powerful approach for enhancing pharmacological potency and selectivity. Nevertheless, concerns about off-target modification motivate the development of reversible cysteine-targeting strategies. Here we show that electron-deficient olefins, including acrylamides, can be tuned to react with cysteine thiols in a rapidly reversible manner. Installation of a nitrile group increased the olefins’ intrinsic reactivity, yet paradoxically eliminated the formation of irreversible adducts. Incorporation of these electrophiles into a noncovalent kinase recognition scaffold produced slowly dissociating, covalent inhibitors of the p90 ribosomal protein S6 kinase, RSK. A cocrystal structure revealed specific noncovalent interactions that stabilize the complex by positioning the electrophilic carbon near the targeted cysteine. Disruption of these interactions by protein unfolding or proteolysis promoted instantaneous cleavage of the covalent bond. Our results establish a chemistry-based framework for engineering sustained covalent inhibition without accumulating permanently modified proteins and peptides.
Cysteine displays rich chemistry through its nucleophilic thiol group. It is also one of the least common amino acids in proteins. Together, these properties make cysteine residues ideal for targeting with covalent drugs, which have the potential to exhibit high levels of target specificity and a prolonged duration of action1-3. Although frequently designed to inactivate conserved, catalytically essential nucleophiles (e.g., in Ser, Thr, and Cys proteases), covalent inhibitors can achieve maximal selectivity among related targets by exploiting the intrinsic nucleophilicity of poorly conserved, noncatalytic cysteines4. This strategy, guided by structural bioinformatics analysis, has led to the design of selective, irreversible inhibitors of protein kinases5-9, and more recently, the NS3/4A serine protease from hepatitis C virus10.
Protein kinases are challenging therapeutic targets from the standpoint of achieving sustained inhibition of the desired kinase without affecting structurally related kinases. A majority of the 518 human kinases have an accessible noncatalytic cysteine within reach of the active site11,12, and at least four cysteine-targeted kinase inhibitors are in clinical trials for advanced cancer indications. They all rely on an acrylamide electrophile to form an irreversible covalent bond with the kinase4. Acrylamide-based kinase inhibitors react irreversibly with glutathione13 and therefore may react with proteins other than the desired target, especially proteins with hyper-reactive cysteines14. Although the risk may be low and more relevant to chronic diseases than advanced cancer, there are currently no preclinical models that can accurately predict the toxicological potential of chemically reactive drugs and drug metabolites15-17. Thus, current drug discovery efforts mostly aim to avoid the formation of irreversible covalent adducts.
Based on these considerations, we sought reversible electrophilic inhibitors that would retain the advantages of covalent cysteine targeting (prolonged duration of action and high selectivity) without the potential liabilities associated with irreversible adduct formation. The few known covalent inhibitors that reversibly target noncatalytic cysteines were discovered by random high-throughput screening18,19, and the chemical basis of their reversibility is not clear. In this study, we elucidate specific structural features underlying reversible thiol addition to electron-deficient olefins and apply these principles to the design of reversible, cysteine-targeted kinase inhibitors.
RESULTS
Reversibility of thiol addition to activated olefins
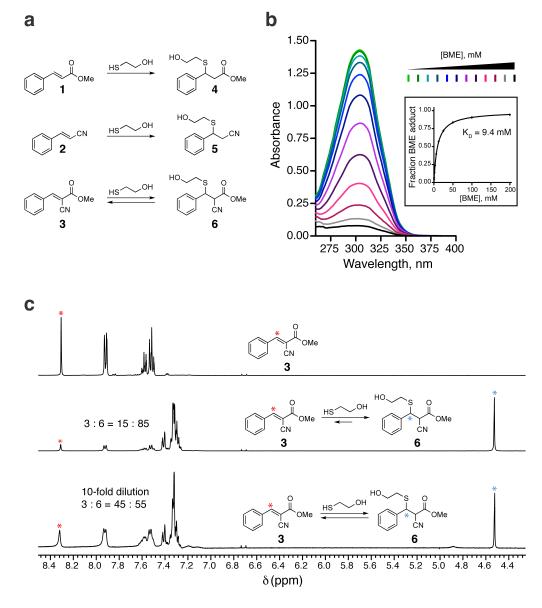
Experiments in the 1960s revealed that simple thiols react instantaneously with 2-cyanoacrylates at physiological pH, but the products could not be isolated or structurally characterized20. A potential explanation for these results is that the reaction, possibly a Michael-type conjugate addition, is a rapid-equilibrium process. To test this hypothesis and define the structural requirements for rapid reversibility, we compared three simple Michael acceptors, activated by a methyl ester (1), a nitrile (2), or both electron-withdrawing groups (3) (Fig. 1a). Reactions of acrylate 1 and acrylonitrile 2 with the model thiol, beta-mercaptoethanol (BME), produced the stable thioether adducts 4 and 5, which were easily isolated and characterized (Supplementary Results, Supplementary Fig. 1). By contrast, when the doubly activated Michael acceptor 3 was treated with BME (Fig. 1a), only the starting cyanoacrylate was recovered. Addition of increasing concentrations of BME caused a stepwise reduction in the prominent UV-visible absorption band of cyanoacrylate 3 (λmax 304 nm), and fitting these titration data provided an apparent equilibrium dissociation constant (KD) of 9.4 mM (Fig. 1b). 1H NMR provided further spectroscopic evidence for the formation of an adduct corresponding to thioether 6, and dilution experiments confirmed that the reaction was rapidly reversible (Fig. 1c). The facile reversion of thioether adduct 6 to the starting cyanoacrylate likely derives from its enhanced kinetic and thermodynamic acidity, estimated21 to be 10–15 orders of magnitude greater than the carbon acidity of stable adducts 4 and 5. Hence, the combined influence of a nitrile and an ester on the acidity of the alpha C–H bond facilitates rapid elimination of thiol adducts at physiological pH, while at the same time accelerating the rate of thiol addition.
Figure 1. Thiol reactivity of electron-deficient olefins.
(a) Conjugate addition reactions of beta-mercaptoethanol (BME) with olefins 1–3. (b) Cyanoacrylate 3 (100 μM) was treated with increasing concentrations of BME and monitored by UV-visible absorption spectroscopy. The inset shows the fraction of BME adduct versus the concentration of BME, from which the equilibrium dissociation constant (KD) was derived. (c) 1H NMR spectra showing reversible addition of BME to cyanoacrylate 3. Cyanoacrylate 3 (90 mM, top spectrum) was treated with 100 mM BME in DMSO-d6:PBS-d (3:1), affording an 15:85 mixture that favored the BME adduct 6 (middle). Upon 10-fold dilution, the equilibrium shifted to the left (bottom). Red and blue asterisks indicate protons used to determine ratios of 3:6.
To test whether this chemistry is applicable to structurally related Michael acceptors, we synthesized five cyanoacrylamides with diverse substituents attached to the electrophilic beta-carbon (7-11, Supplementary Fig. 2). We included entacapone, a cyanoacrylamide-based drug used for the treatment of Parkinson’s disease; to our knowledge, the reactivity of entacapone toward thiols has not been reported previously. Similar to cyanoacrylate 3, the cyanoacrylamides reacted with millimolar concentrations of BME in a manner that was reversed within seconds upon dilution (Supplementary Fig. 2). Depending on the beta-substituent, the equilibrium dissociation constants ranged from 0.2 to 33 mM, corresponding to standard free energy changes (ΔGo) of –2 to –5 kcal/mol. The ability of beta-substituted cyanoacrylamide derivatives, including a widely used drug, to form rapidly reversible adducts with thiols suggested an attractive approach for targeting noncatalytic cysteines in small-molecule binding sites, without the use of irreversible electrophiles.
Rational design of reversible covalent kinase inhibitors
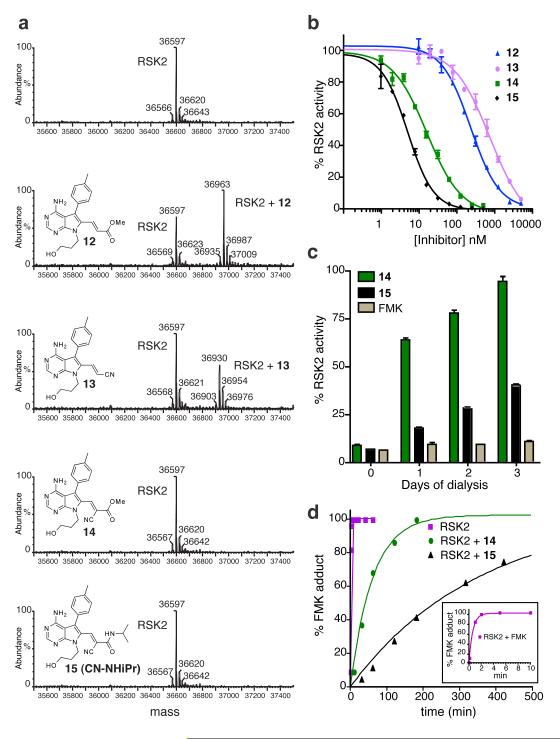
We prepared a series of electrophilic pyrrolopyrimidines (12-15; Fig. 2a), analogous to the model Michael acceptors in Figure 1, and compared their biochemical activities against the C-terminal kinase domain (CTD) of p90 ribosomal protein S6 kinase, RSK2. The electrophilic beta-carbon was designed to be proximal to Cys436 in the RSK2 active site based on the predicted binding orientation of the pyrrolopyrimidine scaffold, similar to the irreversible fluoromethylketone-based inhibitor developed previously (‘FMK’; see Supplementary Fig. 4 for structure) 6,22. Treatment of the RSK2-CTD with five equivalents of acrylate 12 or acrylonitrile 13 led to the slow formation of a covalent adduct, as revealed by liquid chromatography–mass spectrometry (LC-MS) (Fig. 2a). By contrast, the doubly activated Michael acceptors 14 and 15 failed to produce adducts detectable by LC-MS (Fig. 2a), despite being more than 200-fold more potent than the singly activated Michael acceptors 12 and 13 in kinase activity assays (Fig. 2b). Mutation of Cys436 to valine conferred ~1000-fold resistance to inhibitors 14 and 15 (Supplementary Fig. 3), suggesting that potent RSK2 inhibition requires covalent bond formation. Cyanoacrylamide 15 reacted rapidly and reversibly with BME and glutathione (KD = 5.1 and 7.3 mM), yet pretreatment with 10 mM glutathione had no effect on its ability to inhibit RSK2 (IC50 = 4 nM; Supplementary Fig. 3). Cyanoacrylamide 15 did not react substantially with lysine or ethanolamine (up to 100 mM, Supplementary Fig. 3).
Figure 2. Sustained, reversible inhibition of RSK2-CTD by doubly activated Michael acceptors.
(a) RSK2-CTD (5 μM) was treated with pyrrolopyrimidines 12–15 (25 μM) for 1 h, followed by LC-MS analysis. Observed molecular masses of unmodified RSK2-CTD and the 1:1 adducts with 12 and 13 are consistent with the predicted values. (b) In vitro kinase assays of RSK2-CTD. Shown are mean values from duplicate measurements (± range). Data were fit with PRISM 4.0 to provide IC50 values (Supplementary Fig. 3a). (c) RSK2-CTD (50 nM) was treated with the indicated inhibitors (1 μM) or DMSO for 1 h at room temp, prior to dialyzing at 4°C. Aliquots were tested daily for kinase activity, normalized to DMSO control (mean ± SD, triplicate values). (d) RSK2-CTD (5 μM) was pretreated with inhibitors 14 or 15 (10 μM) for 1 h, followed by addition of excess FMK (100 μM final). At each time point, an aliquot was removed and quenched by diluting 1:1 into 0.4% formic acid. Relative amounts of RSK2-CTD and the FMK adduct were quantified by LC-MS. Progress curve fits to a single exponential were used to estimate dissociation half-times of 14 and 15. Inset: progress curve detail showing reaction of RSK2-CTD with 100 μM FMK.
RSK2-CTD kinase activity recovered from inhibition by cyanoacrylate 14 after dialysis for three days at 4°C, whereas recovery from cyanoacrylamide 15 was much slower under these conditions (Fig. 2c). To estimate their biochemical dissociation rates, we developed a kinetic trapping assay in which RSK2-CTD was first saturated with a reversible inhibitor, followed by treatment with a large excess of FMK. In the absence of inhibitor pretreatment, FMK (100 μM) reacted with Cys436 of RSK2-CTD to form a 1:1 covalent adduct (t1/2 < 1 min), as determined by MS analysis (Supplementary Fig. 4). Pretreatment of RSK2-CTD (5 μM) with cyanoacrylate 14 or cyanoacrylamide 15 (10 μM) caused a dramatic reduction in the rate of FMK labeling (Fig. 2d), from which we could estimate dissociation half-times of 42 and 245 min, respectively. Thus, inhibition of RSK2-CTD by cyanoacrylamide 15 is long-lived yet fully reversible, with complete dissociation occurring on a time scale of several hours.
We selected the N-isopropyl cyanoacrylamide 15 (hereafter referred to as ‘CN-NHiPr’) for further biochemical and cellular experiments due to its greater potency and slower off-rate compared to cyanoacrylate 14. Kinase profiling23 revealed that CN-NHiPr is highly selective for the C-terminal kinase domains of RSK1 and RSK4 (RSK2-CTD was not on the panel of profiled kinases). Besides RSK1/4-CTD, only 6 of the 442 kinases profiled showed greater than 70% inhibition by 1 μM CN-NHiPr relative to the DMSO control (Supplementary Table 1). Subsequent KD determinations demonstrated that CN-NHiPr bound RSK1-CTD (KD = 540 pM) with ~80-fold higher affinity than MAP3K1 and at least 400-fold higher affinity than the remaining five kinases (STK16, RIPK2, RET, MEK5, and PDGFRB) (Supplementary Fig. 5).
Sustained, quasi-irreversible occupancy of RSK in cells
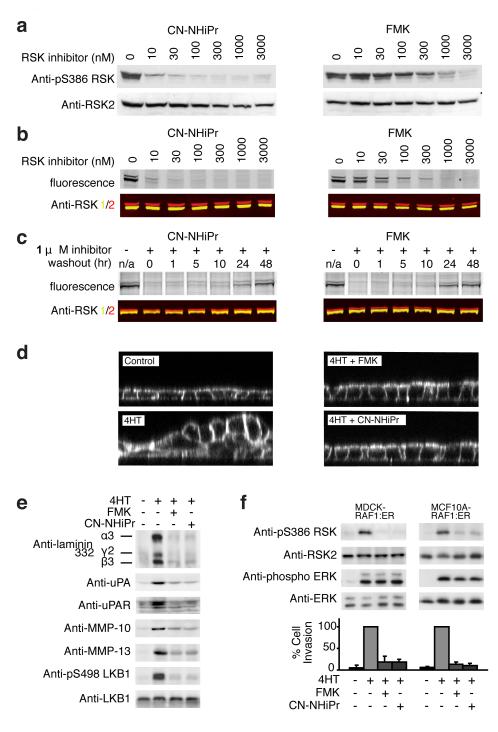
We compared the cellular activity of CN-NHiPr with FMK, previously shown to be an irreversible inhibitor of endogenous RSK1 and RSK26,22. Treatment of MDA-MB-231 breast cancer cells with CN-NHiPr for 2 h led to potent inhibition of RSK2-CTD-mediated autophosphorylation of Ser386 (Fig. 3a, EC50 < 10 nM), a key regulatory site for the N-terminal effector kinase domain24. With a similar dose response, CN-NHiPr prevented an irreversible fluorescent probe, FMK-BODIPY22, from labeling RSK1/2 in cells (Fig. 3b). Despite its irreversible binding mode, FMK was less potent than CN-NHiPr in both the cellular occupancy and autophosphorylation assays, suggesting that CN-NHiPr enters cells and binds to endogenous RSK1/2 with faster kinetics. Washout experiments revealed a striking duration of RSK1/2 occupancy by CN-NHiPr, indistinguishable from FMK. After washout of FMK or CN-NHiPr, reappearance of inhibitor-free RSK1/2 required at least 48 h, presumably the time required for RSK1/2 resynthesis (Fig. 3c). Similar to FMK, CN-NHiPr abolished RAF-induced epithelial cell migration and invasion (Fig. 3d–f), consistent with the recently established role of RSK signaling in these processes25-27.
Figure 3. Sustained inhibition of cellular RSK1/2 by CN-NHiPr.
(a) MDA-MB-231 cells were treated with CN-NHiPr or FMK. Cell lysates were immunoblotted for pS386-RSK or RSK2. (b) MDA-MB-231 cells were treated with CN-NHiPr or FMK, followed by FMK-BODIPY (3 μM). Cell lysates were resolved by SDS-PAGE and detected by in-gel fluorescence scanning or immunoblotting for RSK1 (yellow) and RSK2 (red). (c) MDA-MB-231 cells were treated with CN-NHiPr or FMK (1 μM). The media was exchanged and cells were harvested at the indicated times after inhibitor washout. Cell lysates were treated with FMK-BODIPY (5 μM). Proteins were resolved on SDS-PAGE and detected by in-gel fluorescence or immunoblotting for RSK1 (yellow) and RSK2 (red). (d) Polarized MDCK-RAF:ER cell monolayers were treated with CN-NHiPr or FMK (2 μM) and stimulated with 4-hydroxytamoxifen (4HT, 1 μM) to activate RAF-dependent epithelial cell multilayering. After 24 h, cells were imaged by confocal fluorescence microscopy (XZ plane). (e) Polarized MDCK-RAF:ER cell monolayers were treated as in d. After 24 h, cell lysates and conditioned media were analyzed by immunoblotting for the indicated proteins. (f) MDCK-RAF:ER and MCF10A-RAF:ER cells were treated with CN-NHiPr or FMK (2 μM) and 4HT (1 μM) as indicated. Cells were then either lysed for immunoblot analysis or trypsinized and subjected to Matrigel invasion assays. After 24 h, cell invasion was quantified and expressed as a percentage of the maximum value (mean ± SD from three experiments).
Cocrystal structure of the RSK2-cyanoacrylate complex
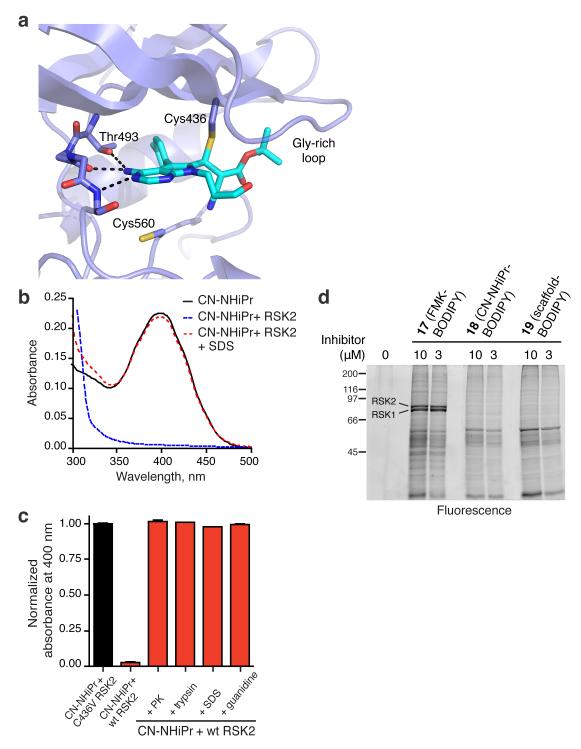
A cocrystal structure provided insight into the molecular basis of sustained RSK2 inhibition by doubly activated Michael acceptors (Fig. 4a). We screened several related cyanoacrylate and cyanoacrylamide inhibitors and obtained the best diffraction data (2.4 Å resolution) with a tert-butyl cyanoacrylate derivative 16 (Supplementary Fig. 3a) that has similar potency and dissociation kinetics (t1/2 = 163 min) as CN-NHiPr. The structure confirms the covalent nature of the complex (Fig. 4a, PDB code 4D9U), with strong electron density connecting Cys436 to the electrophilic beta-carbon of the cyanoacrylate (Supplementary Fig. 6a). A second cysteine (Cys560) within ~7 Å of the electrophilic carbon is incapable of forming a stable covalent bond, as indicated by the ~1000-fold decrease in potency of CN-NHiPr toward C436V RSK2 (Supplementary Fig. 3). Hydrogen bonds between the pyrrolopyrimidine scaffold, the RSK2 hinge region, and the side chain of Thr493 position the electrophilic beta-carbon beneath Cys436 and are likely essential for driving covalent bond formation (Fig. 4a). An additional anchor is provided by the p-tolyl group, which packs against the gatekeeper side chain (Thr493) and extends into a hydrophobic pocket. Mutation of Thr493 to Met in RSK2-CTD conferred ~1000-fold resistance to CN-NHiPr (Supplementary Fig. 6b). RSK3-CTD and the related kinase domains of MSK1/2 have a Met gatekeeper, explaining their insensitivity to CN-NHiPr (Supplementary Table 1).
Figure 4. Specific noncovalent interactions drive covalent bond formation.
(a) Co-crystal structure of RSK2-CTD bound to tert-butyl cyanoacrylate 16. Ribbon representation of RSK2-CTD with cyanoacrylate 16 in green highlights hydrogen bonds to the hinge and the gatekeeper, Thr493. Note the close proximity of Cys560, which does not form a covalent bond with the electrophilic beta-carbon. (b) UV-visible spectra of CN-NHiPr showing the 400 nm cyanoacrylamide absorption peak. CN-NHiPr (20 μM, black curve) was treated with wild-type RSK2-CTD (25 μM), resulting in a loss of the cyanoacrylamide peak (blue curve). Addition of SDS (2% final) caused instantaneous recovery of the cyanoacrylamide (red curve). (c) Effect of protein unfolding (2% SDS; 3 M guanidine HCl) and proteolysis (proteinase K, PK; trypsin) on the covalent complex derived from CN-NHiPr (20 μM) and RSK2-CTD (25 μM). Recovery of free CN-NHiPr was determined by quantifying the absorbance at 400 nm (triplicate measurements, mean ± SD). C436V RSK2-CTD (25 μM) had no effect on the CN-NHiPr absorption peak. (d) Modification of cellular proteins by BODIPY conjugates of FMK (17), CN-NHiPr (18), and the pyrrolopyrimidine scaffold (19) (see Supplementary Fig. 8 for structures). MDA-MB-231 cells were treated with the indicated BODIPY conjugates. Cell lysates were resolved by SDS-PAGE and BODIPY adducts detected by in-gel fluorescence scanning.
Protein unfolding promotes covalent bond dissociation
If the combined nonbonded interactions between RSK2 and the pyrrolopyrimidine are essential for stabilizing the covalent bond, disruption of the kinase domain’s tertiary fold should promote rapid elimination of Cys436 and regeneration of the original Michael acceptor. We tested this hypothesis by monitoring covalent bond formation in solution with UV-visible spectroscopy. Treatment of CN-NHiPr (20 μM in pH 7.5 buffer) with a slight excess of RSK2-CTD (25 μM) caused disappearance of the strong 400 nm absorption peak (Fig. 4b,c), consistent with nucleophilic attack on the cyanoacrylamide. Mutation of Cys436 abolished this effect (Fig. 4c), demonstrating that none of the remaining cysteines (including Cys560 in the ATP binding site) detectably react with CN-NHiPr, even at millimolar concentrations (Supplementary Fig. 7). When the covalent RSK2-CTD-CN-NHiPr complex was unfolded by the addition of sodium dodecylsulfate or guanidine, the cyanoacrylamide absorption peak reappeared within seconds (Fig. 4b,c), and LC-MS analysis indicated quantitative recovery of CN-NHiPr (Supplementary Fig. 7). Proteolytic digestion of the complex with trypsin or proteinase K also promoted complete reversal of the covalent bond (Fig. 4c), suggesting that cyanoacrylamide-modified peptides derived from cellular RSK turnover would be short-lived.
We have shown that cyanoacrylamides react reversibly with model thiols and recombinant RSK2-CTD. However, formation of irreversible adducts with endogenous full-length RSK or other cellular proteins remained possible, especially given the apparent irreversible binding to RSK1/2 (resistant to washout) in cells (Fig. 3c). We addressed this possibility by treating cells with fluorescent BODIPY conjugates derived from FMK, CN-NHiPr, and the pyrrolopyrimidine scaffold as a non-electrophilic control (see Supplementary Fig. 8 for chemical structures). Analysis of cell lysates by SDS-PAGE revealed several prominent bands labeled by the irreversible probe, FMK-BODIPY, including endogenous RSK1 and RSK222 (Fig. 4d). By contrast, BODIPY conjugates of CN-NHiPr and the non-electrophilic scaffold failed to label RSK1/2. Instead, both BODIPY conjugates bound weakly to an identical subset of denatured proteins, presumably in a noncovalent manner. We conclude that specific interactions provided by the folded kinase domain, in concert with a precisely positioned cysteine, are required to cooperatively stabilize a covalent complex with CN-NHiPr. The lack of irreversible adduct formation by CN-NHiPr in a cellular context is remarkable given that it is more than 50,000 times more reactive toward thiols than FMK (reaction with 10 mM glutathione: CN-NHiPr t1/2 < 1 sec; FMK t1/2 = 16 h).
DISCUSSION
In this study, we report a novel approach for targeting noncatalytic cysteines with reversible covalent inhibitors. The predominant strategy for targeting noncatalytic cysteines, exemplified by several acrylamide-based kinase inhibitors in clinical trials, has been to minimize the intrinsic chemical reactivity of the electrophile as much as possible4. This strategy derives from the chemically intuitive hypothesis that attenuated electrophiles have a reduced likelihood of reacting irreversibly with off-target proteins. Our results challenge this notion. Although attenuated Michael acceptors such as acrylamides may react slowly with most cysteine residues, the probability of modifying a hyper-reactive cysteine (comprising up to ~10% of solvent-exposed cysteines14) is unpredictable; such off-target reactions are clearly undesirable.
Our chemical and biochemical experiments demonstrate that addition of a nitrile group (with a molecular mass of only 26 Da) converts irreversible Michael acceptors (e.g., acrylate or acrylamide) into electrophiles with unanticipated properties. The resulting electrophiles (e.g., cyanoacrylate or cyanoacrylamide) react with cysteine thiols under physiological conditions in a manner that is energetically favorable yet rapidly reversible, thus minimizing the chance of producing irreversibly modified peptides. Adding the nitrile group to the alpha-carbon of an acrylamide increases the susceptibility of the beta-carbon to nucleophilic attack but also stabilizes the resultant carbanion. This leads to a kinetic regime in which thiol addition and elimination proceed on a subsecond-second time scale, a surprising result that, to our knowledge, has not been demonstrated previously. Rapid thiol-addition/elimination chemistry appears to be a general property of cyanoacrylamides, and in preliminary work, extends to some, but not all, combinations of two electron-withdrawing groups in doubly activated Michael acceptors. We envision that the reversible covalent targeting strategy described herein can be applied to any kinase that has an exposed cysteine within striking distance of the active site, provided that the geometric requirements for covalent bond formation are satisfied with minimal strain. More generally, by using structural bioinformatics to identify solvent-exposed cysteines near potential small-molecule binding sites, it may be possible to apply this strategy to additional target classes outside of the protein kinase family.
METHODS
Chemicals
The following chemicals were commercially obtained and used without further purification: Methyl trans-cinnamate (1), 99% pure, Sigma-Aldrich; cinnamonitrile (2) 97% pure, Sigma-Aldrich; Entacapone, 99% pure, AK Scientific; Phorbol 12-myristate 13-acetate (PMA) >99% pure, Sigma-Aldrich; and (Z)-4-Hydroxytamoxifen (4HT) >97% pure, Calbiochem.
Inhibitors
Compounds 3-16 and BODIPY conjugates 17-19 were synthesized and characterized as described in the Supplementary Methods. FMK was synthesized as previously described6 and the characterization and purity matched published standards.
Protein Purification
RSK2-CTD was expressed and purified as previously described28. Briefly, wild type and mutant RSK2-CTDs were expressed in E.coli from pET-46 EK/LIK His6 fusion vectors and purified by Ni/NTA affinity chromatography in 50 mM Tris pH 8.0, 0.5 M NaCl and 10 – 500 mM imidazole gradient, followed by cleavage of the His6-tag and a second purification with S75 size exclusion column.
Protein kinase activity assays
RSK2-CTD was assayed for kinase activity as described6,22. Detailed methods describing the determination of inhibitor IC50 values for RSK2-CTD, as well as kinase activity recovery after inhibitor dialysis are provided in the Supplementary Methods.
Covalent modification of RSK2-CTD
LC-MS was used to detect covalent adducts between the various pyrrolopyrimidine electrophilic derivatives and RSK2-CTD. Purified RSK2-CTD (5 μM) was incubated with the various inhibitors (25 μM) for 1 h at room temp and was analyzed by liquid chromatography and in-line ESI mass spectrometry as described in the Supplementary Methods.
UV-visible spectroscopic assays
Detailed methods for determining the dissociation constants of electrophile/thiol adducts are provided in the Supplementary Methods. Briefly, electrophilic compounds (100–200 μM in PBS, pH 7.4) were treated with increasing concentrations of BME or GSH prior to acquiring absorption spectra. The extent of electrophile consumption was determined by monitoring the disappearance of the absorbance at λmax.
Cellular assays
Cellular RSK inhibition and occupancy using FMK-BODIPY was determined as described22. MDA-MB231 cells were grown until confluence in the presence of DMEM supplemented with 10% serum. Prior to inhibitor treatment, the media was exchanged to serum free DMEM and the cells were treated for 2 h with the various inhibitors, lysed and analyzed by various methods as described in the Supplementary Methods. Multilayering and Matrigel invasion assays using MDCK-RAF:ER cells were performed as described25.
Crystal structure determination and refinement
Cocrystals of tert-butyl cyanoacrylate 16 bound to RSK2-CTD were obtained in 0.1 M Tris pH 8.5, 25% PEG 3350 (PDB code 4D9U). Details about data collection, structure determination, and refinement are described in the Supplementary Methods. Briefly, purified RSK2-CTD was concentrated to between 5 mg/mL and 10 mg/mL in buffer (20 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT) and incubated with one molar equivalent of cyanoacrylate 16. Hanging drops of 1 μL protein – cyanoacrylate 16 and 1 μL precipitant solution (0.1 M Tris pH 8.5, 25% PEG 3350) were used to obtain crystals. Crystals were cryo-protected in mother liquor with 30% ethylene glycol and flash frozen in liquid nitrogen prior to crystallographic data acquisition. The data sets were processed using XDS and the structure was solved by molecular replacement.
Supplementary Material
Acknowledgements
We thank David King (HHMI Mass Spectrometry Laboratory) for protein mass spectrometry expertise, members of the Taunton lab for insight, and the staff of ALS Beamline 8.3.1 for help with data collection. This work was supported by grants from the National Institutes of Health (GM071434 to J.T., CA020535 and K99CA149088 to M.P., F32GM087052 to J.M.), the Leukemia and Lymphoma Society (5416-7 to M.P.), and the California Tobacco Related Disease Research Program (19FT-0091 to S.K.). We acknowledge the UCSF Mass Spectrometry Facility (supported by NIH grant P41RR001614).
Footnotes
Author Contributions. J.T. conceived and directed the study. I.M.S., S.K., M.S.C., R.L.M., J.M.M., and R.M.M. synthesized compounds, designed and executed chemical, biochemical and cellular experiments, and analyzed data. K.D. and M.F. designed and executed the cellular multilayering and invasion experiments. M.A.P. solved and refined the cocrystal structure. J.T., I.M.S., and S.K. wrote the manuscript with contributions from all other authors.
Competing Financial Interest. J.T., I.M.S., S.K., M.S.C., R.L.M., J.M.M., and R.M.M. are co-inventors on a patent application covering the inhibitors described in this paper.
References
- 1.Copeland RA, Pompliano DL, Meek TD. Opinion - Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.Potashman MH, Duggan ME. Covalent Modifiers: An Orthogonal Approach to Drug Design. J Med Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 3.Smith AJT, Zhang XY, Leach AG, Houk KN. Beyond Picomolar Affinities: Quantitative Aspects of Noncovalent and Covalent Binding of Drugs to Proteins. J Med Chem. 2009;52:225–233. doi: 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 5.Fry DW, et al. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. P Natl Acad Sci USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honigberg LA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. P Natl Acad Sci USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou WJ, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou WJ, et al. A Structure-Guided Approach to Creating Covalent FGFR Inhibitors. Chem Biol. 2010;17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagel M, et al. Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine. Nat Chem Biol. 2011;7:22–24. doi: 10.1038/nchembio.492. [DOI] [PubMed] [Google Scholar]
- 11.Leproult E, Barluenga S, Moras D, Wurtz JM, Winssinger N. Cysteine Mapping in Conformationally Distinct Kinase Nucleotide Binding Sites: Application to the Design of Selective Covalent Inhibitors. J Med Chem. 2011;54:1347–1355. doi: 10.1021/jm101396q. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JM, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 13.Wissner A, et al. Synthesis and structure-activity relationships of 6,7-disubstituted 4-anilinoquinoline-3-carbonitriles. The design of an orally active, irreversible inhibitor of the tyrosine kinase activity of the epidermal growth factor receptor (EGFR) and the human epidermal growth factor receptor-2 (HER-2) J Med Chem. 2003;46:49–63. doi: 10.1021/jm020241c. [DOI] [PubMed] [Google Scholar]
- 14.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–797. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uetrecht J. Idiosyncratic drug reactions: Past, present, and future. Chem Res Toxicol. 2008;21:84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- 16.Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. Drug-protein adducts: An industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 17.Park BK, et al. Managing the challenge of chemically reactive metabolites in drug development. Nat Rev Drug Discov. 2011;10:292–306. doi: 10.1038/nrd3408. [DOI] [PubMed] [Google Scholar]
- 18.Lee G, et al. Novel inhibitors of hepatitis C virus RNA-dependent RNA polymerases. J Mol Biol. 2006;357:1051–1057. doi: 10.1016/j.jmb.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Patch RJ, et al. Identification of Diaryl Ether-Based Ligands for Estrogen-Related Receptor alpha as Potential Antidiabetic Agents. J Med Chem. 2011;54:788–808. doi: 10.1021/jm101063h. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard RB, Lough CE, Currie DJ, Holmes HL. Equilibrium Reactions of N-Butanethiol with Some Conjugated Heteroenoid Compounds. Can J Chemistry. 1968;46:775–781. [Google Scholar]
- 21.Pearson RG, Dillon RL. Rates of Ionization of Pseudo Acids. IV. Relation between Rates and Equilibria. J Am Chem Soc. 1953;75:2439–2443. [Google Scholar]
- 22.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 24.Frodin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 25.Doehn U, et al. RSK Is a Principal Effector of the RAS-ERK Pathway for Eliciting a Coordinate Promotile/Invasive Gene Program and Phenotype in Epithelial Cells. Mol Cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SM, et al. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120:1165–1177. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smolen GA, et al. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Gene Dev. 2010;24:2654–2665. doi: 10.1101/gad.1989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malakhova M, et al. Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nat Struct Mol Biol. 2008;15:112–113. doi: 10.1038/nsmb1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.