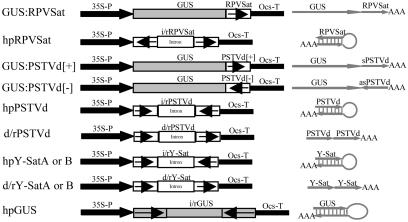
Fig. 1.
Schematic diagrams of transgene constructs used in tobacco and/or tomato transformation. (Right) The predicted structure of RNA transcripts from these transgenes. RPVSat is a full-length (322-nt) sequence of the cereal yellow dwarf virus RPV satellite. The PSTVd sequence in GUS:PSTVd[+] and GUS:PSTVd[-] contains several minor sequence mutations (arising from cloning) compared with the 359-nt wild-type PSTVd-RG1 sequence (deletions of G168 and C318G319 and single-nucleotide substitutions of C185→T and C231→G), but these mutations do not cause significant alteration to the predicted rod-like structure of the viroid RNA. The sense PSTVd sequence in hpPSTVd starts at nucleotide 16 and ends at nucleotide 355 of the RG1 strain, with one nucleotide substitution of G281→A, and the antisense sequence is the same as that in the GUS:PSTVd fusion constructs. There are two versions (A and B) for the hpY-Sat and d/rY-Sat constructs; the Y-SatA sequence has the size of a wild-type Y-Sat RNA (369 nt) but with two single-nucleotide substitutions of A293→G and A308→G, and Y-SatB has a 5-nt deletion (nucleotides 192-196), which constitutes part of the yellow symptom domain, plus four single-nucleotide substitutions of G80→A, G128→ C, G204→T, and A214→T. The hpGUS sequence is the same as that described in ref. 18. 35S-P, cauliflower mosaic virus 35S promoter; Ocs-T, 3′ region of Agrobacterium octopine synthase gene; i/r, inverted-repeat sequences.