Abstract
In Saccharomyces cerevisiae, repair of insertion/deletion loops is carried out by Msh2-Msh3-mediated mismatch repair (MMR). Msh2-Msh3 is also required for 3’ non-homologous tail removal (3’NHTR) in double-strand break repair. In both pathways, Msh2-Msh3 binds double-strand/single-strand junctions and initiates repair in an ATP-dependent manner. However, the kinetics of the two processes appear different; MMR is likely rapid in order to coordinate with the replication fork, whereas 3’ NHTR has been shown to be a slower process. To understand the molecular requirements in both repair pathways, we performed an in vivo analysis of well conserved residues in Msh3 that are hypothesized to be required for MMR and/or 3’NHTR. These residues are predicted to be involved in either communication between the DNA-binding and ATPase domains within the complex or nucleotide binding and/or exchange within Msh2-Msh3. We identified a set of aromatic residues within the FLY motif of the predicted Msh3 nucleotide binding pocket that are essential for Msh2-Msh3-mediated MMR but are largely dispensable for 3’NHTR. In contrast, mutations in other regions gave similar phenotypes in both assays. Based on these results, we suggest the two pathways have distinct requirements with respect to the position of the bound ATP within Msh3. We propose that the differences are related, at least in part, to the kinetics of each pathway. Proper binding and positioning of ATP is required to induce rapid conformational changes at the replication fork, but is less important when more time is available for repair, as in 3’ NHTR.
Introduction
The DNA mismatch repair (MMR) system is an evolutionarily conserved DNA repair pathway that is critical for maintaining genome stability, particularly through the recognition and elimination of errors that occur during DNA synthesis.1–3 Nucleotide misincorporation or DNA slippage events are recognized as mismatches by MutS homologues, or Msh proteins. In contrast to bacteria that contain a single MutS protein, in most eukaryotes, including Saccharomyces cerevisiae and humans, two heterodimeric complexes, Msh2-Msh3 and Msh2-Msh6 act to recognize mismatches and insertion/deletion loops (IDLs). Msh2-Msh3 recognizes, binds and directs repair of IDLs up to 17 nucleotides long4–5 (Fig. 1), while Msh2-Msh6 targets mispairs and IDLs of 1–2 nucleotides. Recently Msh2-Msh3 was also shown to have affinity for some mispairs, especially C-C mispairs.6 Following Msh complex binding to a mispair or loop, a MutL homologue (Mlh) complex is recruited to form a ternary complex that is thought to activate the downstream events in MMR, which include strand unwinding and excision of the nascent strand to remove the error and resynthesis of the DNA. The importance of MMR is reflected in the fact that defective MMR in humans has been associated with hereditary non-polyposis colorectal cancer (HNPCC), a dominant cancer syndrome that results in increased susceptibility to a number of cancers and early age of disease onset.7
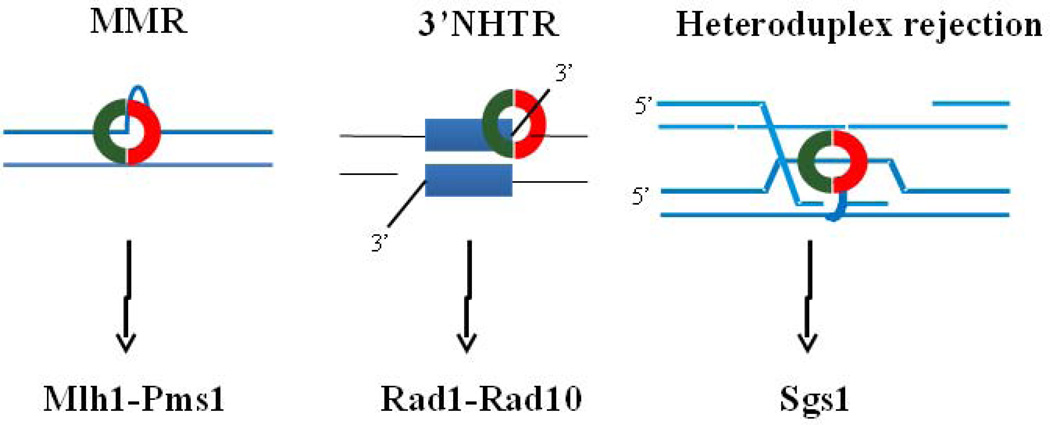
Figure 1.
Msh2-Msh3 is involved in different types of DNA repair. Left panel: Msh2-Msh3 (red and green ring) binds insertion/deletion loops and targets them for mismatch repair, which requires the downstream complex Mlh1-Pms1.23Middle panel: Msh2-Msh3 is required for 3’ non-homologous tail removal. In gene conversion models, a 3’ tail invades a donor strand that has non-homology at the end of the homologous sequence. A stable recombination intermediate that contains 3’ non-homologous tails forms and the tails must be removed to allow DNA synthesis and repair. Rad1-Rad10 cleaves the 3’ non-homologous tails. 54 Right panel: Msh2-Msh3 binds loops that form at homeologous sequences during strand invasion and recruits the helicase Sgs1 to unwind the recombination intermediate, leading to heteroduplex rejection. 43
In vivo and in vitro work with wild-type and mutant MutS and Msh2-Msh6 has demonstrated that the coordination of DNA binding, nucleotide binding and ATP hydrolysis is essential for MMR8–22 and is coupled to specific mispair-binding1; 23; nucleotide binding and hydrolysis are dispensable for mispair-binding but are essential for repair. But much less is known about these requirements within Msh2-Msh3. Indeed the details with respect to nucleotide binding, exchange and hydrolysis appear to differ between Msh2-Msh6 and Msh2-Msh3.24–25 Similarly, mutational and crystallographic evidence indicates that the mode of DNA substrate recognition by Msh2-Msh3 is quite distinct from that of MutS and Msh2-Msh6.26–30 Nonetheless, a similar sequence of events has been proposed for Msh2-Msh3.24 Specific DNA-binding by MutS or Msh2-Msh6 induces a sequence of conformational changes within the complex, which increases the affinity of the Msh complex for ATP and initiates nucleotide binding.31–32 ATP-binding acts as a molecular switch, inducing additional conformational changes that, among other things, lead to reduced affinity of the Msh complex for its specific DNA substrate. The altered conformations when ATP is bound are also required for interactions with Mlh complexes9; 33 and for the transition to a sliding clamp conformation that can move away from the mispair or loop.15; 17; 34 This movement away has been proposed to allow multiple loadings of Msh complexes to amplify the signal for MMR.35–36 ATP hydrolysis is not required for movement away from the mispair or loop; ATPγS also supports formation of a sliding clamp.15; 34; 37 Instead, ATP hydrolysis is thought to be important for Msh complex turnover, allowing it to dissociate and subsequently bind specific substrates again, in the ADP-bound form. This model invokes careful coordination of DNA binding with nucleotide binding and hydrolysis to allow proper regulation of the steps leading to MMR.
Msh2-Msh3’s IDL binding activity is also reduced in the presence of ATP.24; 26; 38–39 Conversely, the affinity of Msh2-Msh3 for nucleotide is reduced in the presence of DNA,24 although ATP hydrolysis is enhanced by DNA binding,24; 30; 38–39 as is nucleotide exchange.24 Analogous to Msh2-Msh6, Msh2-Msh3 has been predicted to form an ATP-dependent molecular switch.24; 39 A model based on in vitro analysis of Msh2-Msh3 predicts that the complex binds at double-strand (ds)/single-stranded (ss) DNA junctions26–27 with Msh2 in an ADP-bound state and no nucleotide bound to Msh3.24 Binding specific substrates promotes nucleotide binding and/or exchange at Msh2 and Msh3.24 The resulting complex has nucleotide bound to both subunits and is converted into a sliding clamp that moves away from the mispair.15; 39 ATP hydrolysis follows and the ADP-Msh2-Msh3 complex can again bind DNA lesions with higher affinity. Notably, recent crystallographic evidence has predicted that there may be important differences between Msh2-Msh3 and other Msh complexes with respect to the coordination of nucleotide binding by Msh3.30 Specifically, Msh3 has a conserved aromatic residue (F1023 in human MSH3 (hMSH3), Y925 in yeast Msh3 (yMsh3); see purple residue in Fig.2a) that is not present in MutS, Msh2 or Msh6. This residue is proposed to provide an additional layer of regulation to the occupancy of the Msh3 nucleotide binding site.30
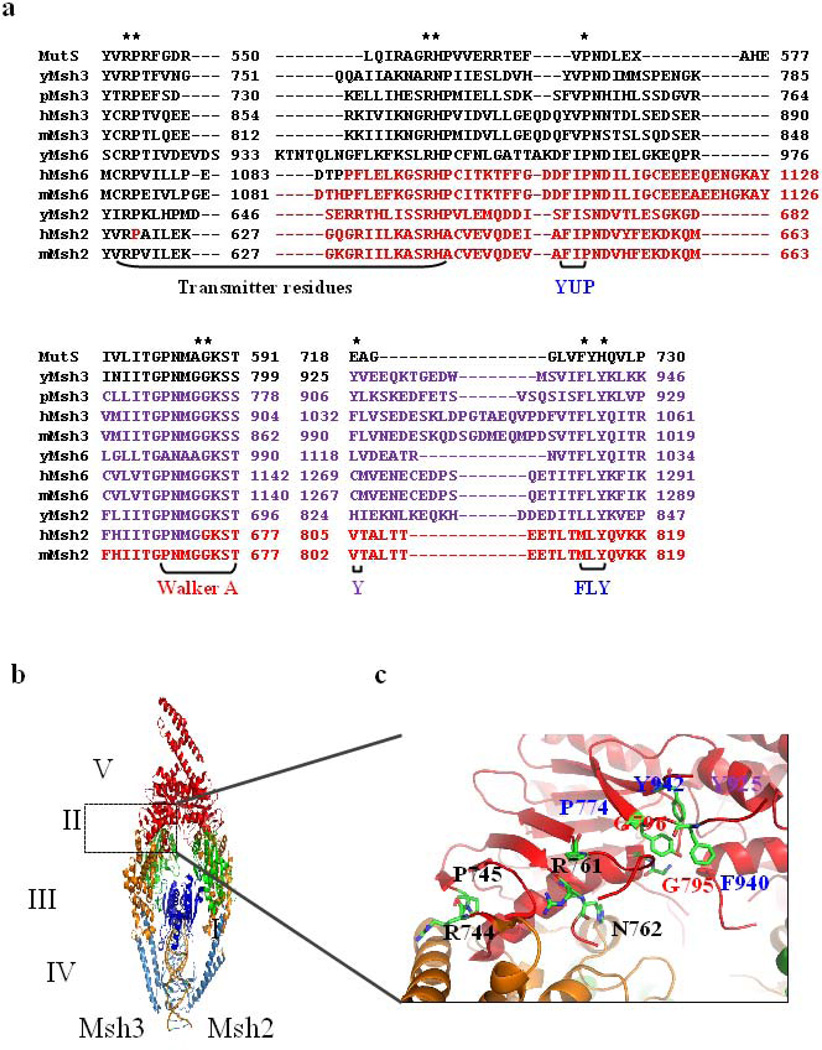
Figure 2. Residues in the transmitter region, Walker A, YUP and FLY motifs.
(a) Sequence alignment of Thermus aquaticus (TAQ) MutS with Msh2, Msh3 and Msh6 sequences from Saccharomyces cerevisiae (y), Schyzosaccharomyces pombe (p), Homo sapiens (h), Mus musculus (m). Residues colored in red represent HNPCC residues in hMsh2 and hMsh6. The regions targeted in this study are indicated by brackets below the alignment. Asterisks indicate residues mutated in this study. (b) The crystal structure of hMsh2-hMsh3 in complex with a four-loop substrate, based on the crystal structure 30. The subunit on the right is Msh2 and Msh3 is on the left. Domains in blue indicate DNA binding domains (domains I & IV), domains in green indicate connector domains (domain II), domains in mustard are the lever domains (domain III), and the ATPase domains (domain V) are in red. The interface of ATPase and lever domains is highlighted by the box. (c) Highlighted in the stick diagrams are residues targeted in this study. They are colored by element (carbon in green and nitrogen in blue). Molecular modeling images were acquired using PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
In addition to MMR, Msh2-Msh3 is involved in other genome stability pathways (Fig. 1). It has been implicated in promoting trinucleotide repeat expansions38; 40–42 and is required during genetic recombination, including the prevention of homeologous recombination, i.e. recombination between divergent sequences.43–44 Msh2-Msh3 is thought to bind loops that can form in strand transfer intermediates, targeting them for unwinding by Sgs1.43; 45–46 Msh2-Msh3 is also involved in the repair of large unpaired loops that can form during meiotic recombination.47–48 In addition to Msh2-Msh3, this repair requires the nucleotide excision repair (NER) structure-specific endonuclease Rad1-Rad10 that cleaves DNA at ds/ss DNA junctions with 3’ ssDNA tails.49–51 Msh2-Msh3 and Rad1-Rad10 are also required in a specialized form of double-strand break repair (DSBR) that involves 3’ non-homologous tail removal (3’ NHTR), for example, single-strand annealing (SSA) and some gene conversion events.52–53 In these pathways, recombination intermediates bearing 3’ non-homologous tails are formed.54 Because DNA polymerases cannot initiate DNA synthesis from an unannealed 3’ tail, the non-homologous tails must be removed in order to complete repair. Msh2-Msh3 is predicted to stabilize the recombination intermediate and the ds/ss DNA junction to facilitate cleavage by Rad1-Rad10;26; 52; 55–57 when the annealed region is greater than 1 kilobase, Msh2-Msh3 is no longer required.52; 57
As in MMR, ATP-binding by Msh2-Msh3 is required for 3’ NHTR12–13 and Msh2-Msh3 dissociates from flap substrates in the presence of ATP in vitro.26 However, it is not known if conformational changes analogous to those in MMR occur upon Msh2-Msh3 binding to a 3’ flap recombination intermediate and, if so, whether they are required for function. In this study, we sought to determine some of the molecular requirements for S. cerevisiae Msh2-Msh3 in MMR and 3’NHTR following DNA binding. To this end we mutated and tested the in vivo function of ten positions in two regions of Msh3 that are predicted to be important for different conformational changes within the Msh2-Msh3 complex: i) the transmitter region, predicted to relay information between the DNA-binding and nucleotide-binding functions of Msh2-Msh3 once bound to a specific DNA substrate and ii) the nucleotide binding pocket, made up of the Walker A motif and the YUP/FLY motifs (identified by sequence homology30), predicted to regulate and coordinate nucleotide binding. These are steps that authorize conformational changes within the complex and interactions with downstream repair components. The mutant msh3 alleles were tested for function in both 3’ NHTR and MMR to assess their effect in each pathway. Most of the alleles had similar effects in both pathways. Importantly, mutations in the FLY region demonstrated a much stronger phenotype in MMR than in 3’ NHTR, indicating that MMR is more stringent in its requirement for nucleotide binding.
Results
In vivo assays for Msh2-Msh3 activity
The ATP binding activity of Msh2-Msh3 is essential for its proper function in MMR and 3’ NHTR.12; 26 However, the requirements, with respect to specific residues within the nucleotide binding pocket, for regulating this activity in Msh2-Msh3 have not been well-studied, particularly in 3’ NHTR. As discussed below, we have performed an in vivo mutational analysis of MSH3, focusing on a portion of the transmitter region between the lever and ATPase domains, the F/YUP and FLY consensus motifs that form part of the nucleotide binding pocket and the Walker A (P-loop) motif of S. cerevisiae Msh3 (Fig. 2). Due to the large number of mutations to be tested, we chose to test the function of the msh3 alleles expressed from low copy number plasmids under the control of the endogenous MSH3 promoter, similar to previous studies.12; 28–29 Therefore we confirmed that the plasmid-borne MSH3 complements the msh3Δ strain for 3’ NHTR.
Previously Lyndaker et al. engineered a yeast strain (EAY1042) that contains double non-homology at the MATa locus and is dependent on Msh2-Msh3 activity for repair following the induction of an HO endonuclease-mediated double strand break (DSB);56 this is a lethal event if left unrepaired. Therefore survival following galactose induction is a measure of DSBR. This strain is also deleted for HMRa, so that repair can only occur via HMLα and therefore involves a mating-type switch from MATa to MATα, which can be monitored by attempting to mate survivors with MATa and MATα strains.56; 58 Only strains of opposite mating type will mate to form diploids. By also measuring switching, we can correct for the possibility that cells survived because they did not suffer a DSB due to incomplete HO induction. Only those cells that have repaired a break will have switched mating type.
In EAY1042 (MSH3), approximately 80% of cells survived induction of a DSB, and 77% of these had undergone a mating-type switch, indicating efficient repair of the break (Table 1). In contrast, only 34% of the msh3Δ derivative of this strain (EAY1118) survived the DSB and 3% of the survivors had switched mating-type (Table 1), indicating very little repair. These results are similar to previous work.56 The low copy number plasmid carrying MSH3 fully complemented the msh3Δ (~75% survival and switching; Table 1), whereas the empty vector did not (36% survival and 8% switching; Table 1).
Table 1.
Percentages of survival of double strand DNA breaks and mating type switching in the presence of msh3 alleles expressed on low copy plasmids.
| Straina | Plasmidb | % Survivalc | % Switchingd | Tetranucleotide repeat | Summaryf | ||
|---|---|---|---|---|---|---|---|
| Mutation ratee (x 10−6) |
Fold change |
3’NHTR | MMR | ||||
| EAY1042 (MSH3) |
- | 79.8 ± 9.5 | 76.8 ± 6.6 | NDg | - | ||
| EAY1118 (msh3Δ) |
- | 35.4 ± 5.0 | 3.3 ± 3.3 | ND | - | ||
| EAY1118 | MSH3 | 74.2 ±1.7 (100%) |
73.3 ± 2.0 (100%) |
8.6 (7.0–12.5) |
1 | ++ | ++ |
| EAY1118 | Empty Vector | 36.1 ± 2.1 (48.7%) |
7.7 ± 3.9 (10.5%) |
228.5 (179.2–273.7) |
26.5 | − | − |
| Transmitter region: | |||||||
| EAY1118 | msh3R744A | 51.1 ± 3.2 (68.9%) | 13.3 ± 2.9 (18.2%) |
77.9 (65.6–99.5) |
9.0 | − | + |
| EAY1118 | msh3R744L | 33.1 ± 3.4 (44.6%) |
5.0 ± 2.9 (6.8%) | ND | - | − | ND |
| EAY1118 | msh3P745A | 75.6 ± 3.2 (100%) |
44.8 ± 3.8 (61.1%) |
ND | - | + | ND |
| EAY1118 | msh3P745L | 29.7 ± 7.2 (40%) | 1.7 ± 1.7 (2.3%) | ND | - | − | ND |
| EAY1118 | msh3R761A | 80.4 ± 2.9 (100%) |
76.2 ± 4.7(100%) | 9.9 (8.0–14.3) |
1.2 | ++ | ++ |
| EAY1118 | msh3R761L | 67.0 ± 2.6 (90.2%) |
66.6 ± 3.4 (90.9%) |
ND | - | ++ | ND |
| EAY1118 | msh3N762A | 73.5 ± 1.7 (99%) | 71.7 ± 1.7 (97.8%) |
ND | - | ++ | ND |
| EAY1118 | msh3N762L | 54.6 ± 3.5 (73.5%) |
66.6 ± 3.4 (90.9%) |
ND | - | ++ | ND |
| Walker A residues: | |||||||
| EAY1118 | msh3G795A | 43.7 ± 8.7 (58.9%) |
36.7 ± 12.01 (50%) |
145.6 (83.0–250.0) |
16.9 | + | − |
| EAY1118 | msh3G795D | 65.3 ± 4.9 (85.6%) |
50.0 ± 8.1 (65.1%) |
ND | - | + | ND |
| EAY1118 | msh3G796A | 43.0 ± 1.9 (58%) | 22.1 ± 10.0 (30%) |
512.0 (352.4–890.0) |
59.3 | + | − |
| EAY1118 | msh3G796D | 39.0 ± 0.5 (52.5%) |
1.7 ± 1.7 (2.3%) | ND | - | − | ND |
| Aromatic residues: | |||||||
| EAY1118 | msh3P774A | 90.0 ± 4.1 (100%) |
76.7 ± 6.7 (100%) |
9.08 (7.5–11.3) |
1.1 | ++ | ++ |
| EAY1118 | msh3P774L | 75.6 ± 6.2 (100%) |
66.7 ± 1.7(91%) | ND | - | ++ | ND |
| EAY1118 | msh3Y925A | 64.7 ± 12 (87.2%) |
61.7 ± 6.0 (84.2%) |
312.7 (290.6–338.6) |
36.3 | ++ | − |
| EAY1118 | msh3F940A | 45.2 ± 3.8 (60.9%) |
30.0 ± 4.3 (40.9%) |
196.0 (117.2–299.5) |
22.7 | + | − |
| EAY1118 | msh3Y942A | 51.3 ± 4.7 (69.1%) |
61.7 ± 4.4 (84.2%) |
120.0 (98.8–169.2) |
13.9 | ++ | − |
Strains carrying a double non-homology at the mating type locus were used to determine Msh2-Msh3-dependent 3’NHTR. Genotypes of the strains are provided in Table S1.
EAY1118 was transformed with low copy plasmids bearing MSH3 or msh3 alleles or with the empty vector to determine 3’NHTR. The plasmids are listed in Table S3. The strain numbers are listed in Table S4.
Percentage cell survival (induced/uninduced) was determined by examining the viability of cells plated after a 45 minute induction of HO expression. The standard error of the mean is indicated. The percentage in parentheses indicated the activity of each allele relative to the activity in the presence of the MSH3 plasmid.
Surviving cells were assayed to determine the percentage that had switched mating type following HO expression and had therefore undergone DSBR (see Materials and Methods). The percentage in parentheses indicated the activity of each allele relative to the activity in the presence of the MSH3 plasmid.
EAY1118 carrying empty vector or MSH3 low copy plasmids was transformed with the tetranucleotide repeat reporter plasmid (pBK1). Mutation rates were determined as described in Materials and Methods. Numbers in parentheses represent 95% confidence intervals.
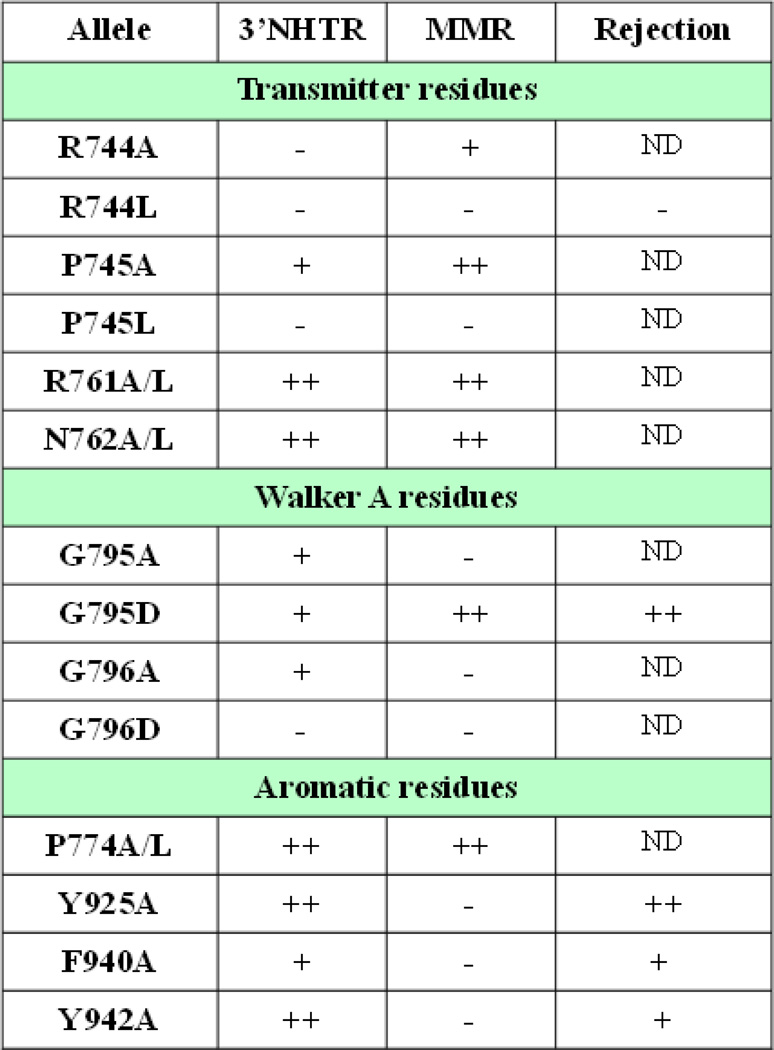
Each allele was characterized as having wild type function (>90% activity; ++), intermediate phenotype (20–90% activity; +) or null phenotype (<20% activity;-) in either 3’ NHTR or MMR.
ND = Not Determined
We constructed seventeen substitution mutations at ten different positions in the transmitter region and the nucleotide binding pocket of MSH3, by site-directed mutagenesis (Table S2 and S3), and performed Western blots using Msh3 antibody to verify the stability of the mutant msh3 proteins. Endogenous Msh3 or Msh3 from msh3Δ cells carrying the low copy MSH3 plasmid was undetectable (data not shown), consistent with the protein being expressed at very low levels.6; 25; 29 The limit of detection of the Msh3 antibody, based on a standard curve using purified protein, was between 50 and 100 ng (data not shown). Therefore we transformed plasmids carrying MSH3 or msh3 alleles under the control of a galactose-inducible promoter, into an msh3Δ strain. From these cleared lysates we were able to detect Msh3 protein. All the mutant msh3 proteins were present at levels comparable to wild-type (Fig. S1) with the exception of msh3P745A, which was present at 10% of the wild-type protein. However, this allele had a very mild phenotype in our assays (see below).
A subset of transmitter residues are important for Msh2-Msh3-mediated 3’ NHTR
The transmitter region at the interface of the lever domain (Domains II and III; Fig. 2) and the ATPase domain (Domain V; Fig. 2) was noted previously10; 32 and was predicted to relay information about the DNA binding status of the complex to the ATPase domains, via conformational changes. Notably, several HNPCC mutations map to this region in Msh2 and Msh632; 59 and sequence alignment of Msh3 orthologs with Msh2 orthologs indicates several highly conserved residues at the interface (Fig. 2). To predict which Msh3 residues might have an altered position following mispair/IDL binding, we overlaid the crystal structure of T. aquaticus MutS (the only Msh complex whose structure has been solved without a DNA substrate) in the absence of DNA with the structure in the presence of mispaired DNA32 to obtain a minimum root mean squared (RMS) value. The validity of the overlay was verified by ensuring the primary sequences of the two structures were aligned. Within the transmitter region, a conserved loop (Fig. 3a, left panel) takes an altered path in the presence of DNA. The side chains of four highly conserved residues in the loop (T.aquaticus MutS R544, P545, R557 and N558 corresponding to yMsh3 R744, P745, R761 and N762; Fig. 2a) are coiled when MutS is bound to a mispair. There were concomitant changes in the F/YUP and FLY motifs of MutS, which form part of the ATP-binding pocket (see below) when MutS bound DNA (Fig. 3a, right panel). We therefore focused on these putative transmitter residues and the nucleotide binding pocket for our in vivo analysis.
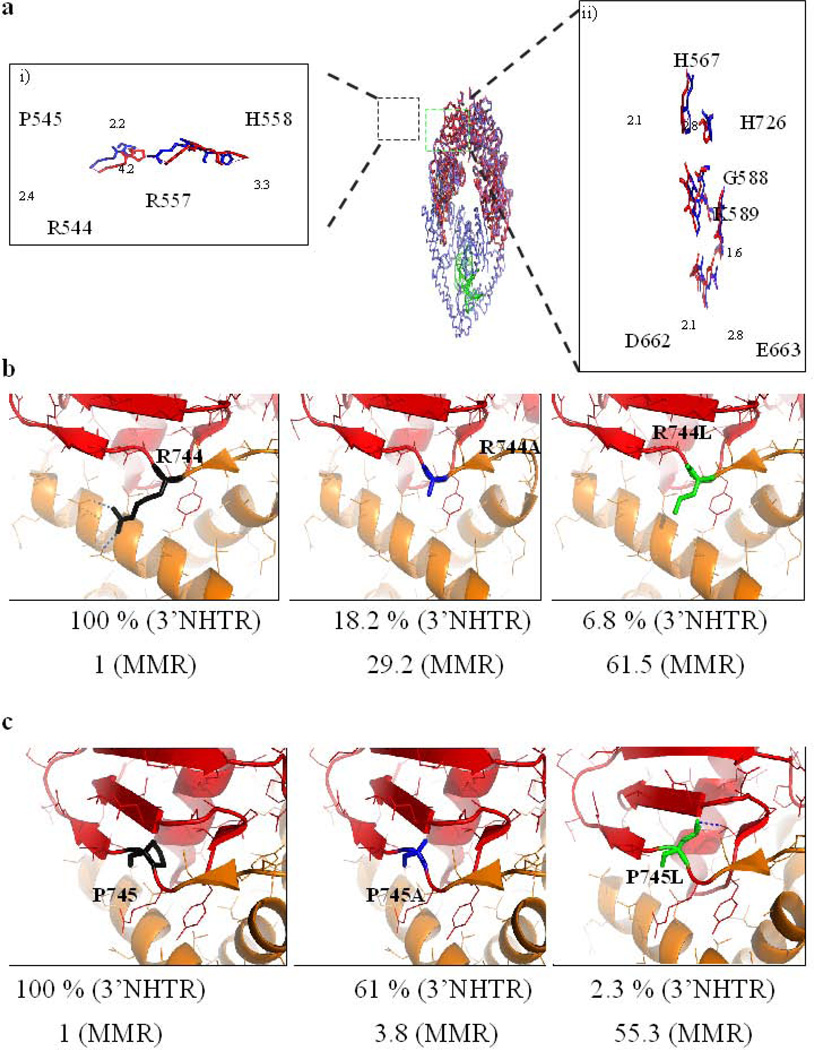
Figure 3. Putative transmitter residues in Msh3.
(a) The crystal structure of TAQ MutS in the absence of DNA (in red, pdb 1EWR) was overlaid with TAQ MutS in the presence of mismatched DNA (in blue, pdb 1EWQ). The left (i) panel zooms in on the four transmitter residues of interest in this study and represents a view of the residues in stick diagrams with the neighboring residues hidden. The image is rotated 30 degrees counter-clockwise around the vertical axis. The R544, P545, R557 and H558 residues of TAQ MutS are analogous to R744, P745, R761 and N762 residues of yMsh3. The dashed lines in magenta represent the translocation (in Å) of the residues in the absence of DNA to a mismatch bound state. The right (ii) panel represents a stick diagram of the active site of ATP hydrolysis of one of the subunits of the TAQ MutS homodimer. The image is rotated 90 degrees clockwise around the vertical axis. Conformations in blue and red represent in the presence and absence of DNA respectively. Dashes in magenta represent distance (in Å) translocated by the residues between the two conformations. (b) R744 of yMsh3 is highlighted in black in the left panel. The R744A (blue) and R744L (green) changes are represented in the center and right panels, respectively. Blue dashes represent polar contacts made by R744 with I764 and E766 . Numbers below each panel represents the activity of Msh2-Msh3 in 3’NHTR relative to wild-type (Table 1) and the fold change in mutation rate of the allele relative to wild-type in the presence of a tetranucleotide repeat reporter plasmid respectively (Table 2). (c) The P745 residue of yMsh3 is represented in black in the left panel. Center and right panels represent P745A and P745L changes respectively. Numbers below each panel represents the activity of Msh2-Msh3 in 3’NHTR relative to wild-type (Table 1) and fold change in mutation rate of the allele relative to wild-type in the presence of a tetranucleotide repeat reporter plasmid respectively (Table 2). The dashed line in blue indicates a potential polar contact between P745L with V817.
We tested the importance of the Msh3 transmitter residues R744, P745, R761, N762 for Msh2-Msh3 function in DSBR in vivo, changing them individually to either alanine or leucine by site-directed mutagenesis (Table 1, Fig. 3). msh3R744A and msh3R744L were defective in DSBR at the MAT locus (18.2% and 6.8% switching, respectively), with msh3R744A exhibiting a milder phenotype in both survival and switching. msh3P745A exhibited wild-type survival but a decrease in switching to 61.8% (compared to 100% with MSH3). This difference may be a result of inefficient HO induction. In contrast, msh3P745L had a null phenotype. This indicates that Msh3 R744 and P745 are important for function in 3’ NHTR. In contrast, msh3R761A, msh3R761L and msh3N762A were wild-type or near wild-type (Table 2). Interestingly, msh3N762L showed reduced survival numbers (73.5%) but all of the survivors had switched mating type. This may be an indication of a defect in the kinetics of repair at the level of 3’ NHTR, i.e. the repair may be slowed down relative to wild-type, resulting in reduced viability, while the mechanism of repair remains intact.
Table 2.
Mutation rates of msh3 alleles.
| Tetranucleotide repeata | Dinucleotide repeatb | ||||
|---|---|---|---|---|---|
| Strain | Allele on plasmid |
Mutation ratec (x10−6) |
Fold change |
Mutation rate (x10−5) |
Fold change |
| FY23 (MSH3) | − | 10.1 (8.3–12.9) |
1 | NDd | - |
| EAY420 (msh3Δ) | − | 559.3 (440–1192.7) |
55.4 | ND | - |
| EAY420 | MSH3 | 6.0 (5.3–6.49) |
1 | 1.4 (1.1–1.9) |
1 |
| EAY420 |
Empty Vector |
346.4 (304–423.5) |
57.7 | 36.16 (27.5–42.6) |
26.3 |
| Transmitter region: | |||||
| EAY420 | msh3R744A | 175.3 (127.7–476.2) |
29.2 | 23.02 (18–26.7) |
16.7 |
| EAY420 | msh3R744L | 369.2 (293.5–409.0) |
61.9 | ND | - |
| EAY420 | msh3P745A | 22.7 (19–36.6) |
3.8 | ND | - |
| EAY420 | msh3P745L | 332.3 (302.7–436.9) |
55.3 | ND | - |
| EAY420 | msh3R761A | 20.45 (18–24.2) |
3.4 | ND | - |
| EAY420 | msh3N762A | 24.2 (18.7–54.9) |
4.0 | ND | - |
| EAY420 | msh3N762L | 26.2 (22.3–33.2) |
4.4 | ND | - |
| Walker A residues: | |||||
| EAY420 | msh3G795A | 208.5 (184.8–236.6) |
34.8 | 42.6 (32.9–47.0 |
30.9 |
| EAY420 | msh3G795D | 17.1 (14.2–23.0) |
2.9 | ND | - |
| EAY420 | msh3G796A | 678.4 (569.6–1087.9) |
113.0 | 228.8 (202.9– 283.5) |
166.2 |
| EAY420 | msh3G796D | 415.8 (289.5–555.2) |
69.3 | ND | - |
| Aromatic residues: | |||||
| EAY420 | msh3P774A | 13.6 (11.2–24.3) |
2.3 | ND | - |
| EAY420 | msh3P774L | 7.4 (6.0–10.1) |
1.2 | ND | - |
| EAY420 | msh3Y925A | 469.5 (444.8–503.3) |
78.3 | ND | - |
| EAY420 | msh3F940A | 343.2 (278.7–644.1) |
57.2 | 29.8 (16.7–59.9) |
21.7 |
| EAY420 | msh3Y942A | 180.1 (148.2–318.7) |
30.0 | 35.2 (32.5–43.5) |
25.1 |
The rate of slippage events within a tetranucleotide repeat were determined by transforming each strain with pBK14, which encodes a (CAGT)16 repeat in frame with the URA3 reporter gene. Slippage events are selected in the presence of 5-FOA.
The rate of slippage events within a dinucleotide repeat were determined by transforming each strain with pSH444, which encodes a (GT)16.5 repeat in frame with the URA3 reporter gene. Slippage events are selected in the presence of 5-FOA.
Mutation rates were determined as described in the Materials and Methods. Numbers in parentheses represent 95% confidence intervals.
ND = Not Determined
Mutations in the Walker A and FLY/YUP motifs exhibited generally mild phenotypes in 3’ NHTR
We focused on two regions of Msh3 that are predicted to be involved in nucleotide binding. The first was the Walker A motif, or P-loop. In ABC ATPases, including Msh proteins, the Walker A motif (consensus sequence GXXXXGK(S/T)) is typically involved in binding the phosphate moieties of ATP and is therefore essential for ATP hydrolysis. The Walker A sequence of Msh3 (and many Msh proteins) is GXXXGGK(S/T). We tested the functional importance of the two glycine residues preceding the lysine in Msh2-Msh3-mediated repair. Both glycines are highly conserved in Msh2 and Msh3, but the first is an alanine in a subset of Msh6 and MutS sequences (Fig. 2a).
Both G795 and G796 were important for Msh2-Msh3 function in 3’ NHTR (Table 1). The presence of either msh3G795A or msh3G795D decreased the efficiency of DSBR (50% switching and 65% switching, respectively), but did not result in a null phenotype. msh3G796A had a stronger phenotype for both survival (58%) and switching (30%) but only msh3G796D exhibited an null phenotype in both survival and switching. Therefore our data indicated both glycines play a role in 3’ NHTR although, with the exception of G796D, the msh3 mutations showed only intermediate phenotypes.
Next, we mutated the so-called F/YUP and FLY motifs of Msh2-Msh3 (and other Msh proteins) that are highly conserved and predicted to form part of the nucleotide binding pocket and regulate the nucleotide occupancy of the Msh complex (Fig. 4a).30 Msh3 is unique among the Msh proteins in having an aromatic residue (Y925 in yMsh3; F1023 in hMSH3) that is predicted to alter the conformation of the FLY motif, causing the phenylalanine (yMsh3 F940) to block the Msh3 nucleotide binding site and prevent nucleotide binding.30 In contrast, this pocket in Msh6 adopts a more open conformation because the FLY motif F is not pushed into the pocket, which is consequently more accessible to nucleotide (Fig. 4b).59
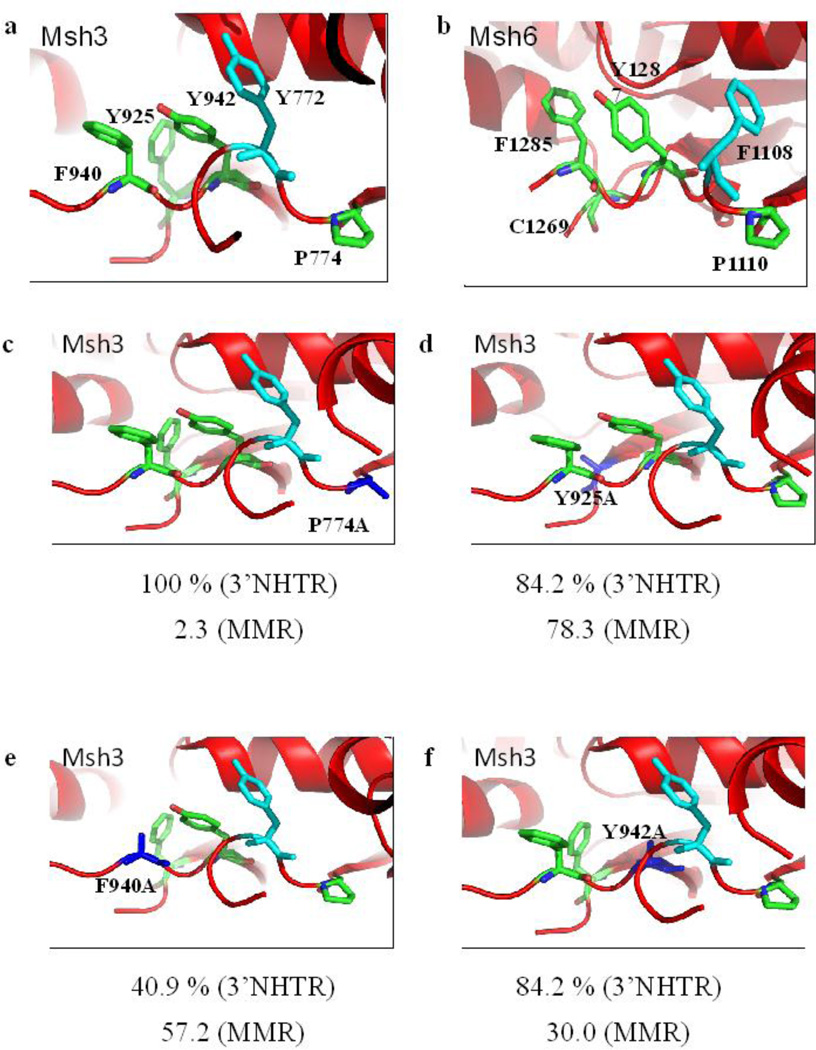
Figure 4. Aromatic residues in the putative nucleotide-binding pocket of Msh3.
(a) The aromatic residues targeted in this study are highlighted in green. The residue in cyan indicates the Y772 residue that forms a nucleotide sandwich with Y942 residue. Y925 pushes F940 into the nucleotide-binding site to block it. P774 is predicted to influence the structure of the adjacent loops, positioning the active site residues. (b) The nucleotide-binding pocket of Msh6 subunit (in hMsh2-hMsh6 complex) is represented. (c) Structural model for P774A mutation is represented. (d) Predicted model for Y925A change is represented. (e) The structural model for F940A mutation is shown. (f) Y942A change is shown. In panels c-f, the changed amino acid is represented in blue. Numbers below each panel represents the activity of Msh2-Msh3 in 3’NHTR relative to wild-type (Table 1) and the fold change in mutation rate of the allele relative to wild-type in the presence of a tetranucleotide repeat reporter plasmid respectively (Table 2).
We changed several residues in these Msh3 consensus motifs to alanine and tested the function of the mutant alleles in 3’ NHTR in vivo (Table 1, Fig. 4). Changing the highly conserved proline of the F/YUP consensus had very little effect on function. Both msh3P774A and msh3P774L were wild-type or near wild-type in 3’ NHTR (Table 1). Surprisingly, alanine mutations at Y925 (the Msh3-specific aromatic residue) and Y942 (of the FLY motif) also had only very mild 3’ NHTR phenotypes, each showing about 84% switching efficiency (Table 1). While msh3F940A (FLY motif) had a more severe defect in 3’ NHTR phenotype than msh3Y925A or msh3Y942A, it nonetheless retained significant activity (~40%) in that pathway.
Mutations in the FLY motif of Msh3 confer differential effects on MMR and 3’ NHTR
The mild DSBR phenotypes of the FLY motif mutations in msh3 were quite unexpected. Therefore we proceeded to characterize these alleles further by testing their MMR phenotypes. To do this, we transformed a subset of the DSBR strains tested above with a tetranucleotide repeat reporter plasmid (pBK1)4 to determine the efficiency of Msh2-Msh3-mediated repair of 4 nucleotide IDLs (Table 1). There were several mutations that exhibited differences in the efficacy of 3’ NHTR versus MMR, particularly within the nucleotide binding pocket. Importantly, the FLY motif alleles exhibited a strong separation of function phenotype. msh3Y925A, msh3F940A and msh3Y942A all exhibited a null phenotype in MMR, consistent with the predicted importance of this motif for MMR,30 while being largely functional in 3’ NHTR (Table 1).
To perform a more extensive analysis of all the msh3 alleles in MMR, we co-transformed the low copy msh3 plasmids with either the tetranucleotide repeat or the dinucleotide repeat reporter plasmid4 into the standard FY23 background that was msh3Δ (EAY420, Table S1) and determined the efficiency of Msh2-Msh3-mediated MMR. The wild-type MSH3 plasmid (pGW2) was able to fully complement the msh3Δ in the repair of plasmid-borne tetranucleotide repeat (pBK14) slippage events (Table 2).
Within the YUP and FLY motifs, the msh3P774A and msh3P774L alleles displayed only a very mild (2-fold) increase in mutation rate (Table 2, Fig. 4), similar to the 3’ NHTR phenotype. However, in striking contrast to their mild effects on 3’ NHTR, altering the aromatic residues of the FLY consensus sequence and the Msh3-specific tyrosine (Y925) conferred a complete loss of Msh2-Msh3-mediated repair. The msh3Y925A allele displayed a 79-fold increase in mutation rate (Fig. 4d). Similarly, msh3F940A conferred a null phenotype in the presence of either the tetranucleotide (58-fold increase) or dinucleotide (22-fold increase) repeat sequences (Table 2, Fig. 4e). And msh3Y942A displayed an elevated (35-fold increase) mutation rate with the tetranucleotide repeat (Fig. 4f) and a null phenotype in the presence of the dinucleotide repeat sequence (Table 2). When overexpressed in an MSH3 background, msh3Y925A and msh3F940A both showed a 12- to 14-fold increase in mutation rate relative to wild-type MSH3 (Table 3). This dominant negative phenotype indicated that these alleles are also able to interfere with the normal MMR function of wild-type Msh2-Msh3 in vivo. These results strongly indicated that MMR and 3’ NHTR have distinct requirements for the FLY motif in particular and perhaps for nucleotide binding in general.
Table 3.
Dominant negative phenotype of msh3 alleles expressed from low copy or overexpression plasmids.
| Low copy plasmida | Overexpression plasmidb | ||||
|---|---|---|---|---|---|
| Strain | Allele | Mutation rate (x10−6) |
Fold change |
Mutation rate (x10−6) |
Fold change |
| FY 23 (MSH3) | MSH3 | 4.4 (3.7–5.2) |
1 | 6.5 (5–10.5) |
1 |
| Transmitter residues: | |||||
| FY 23 | msh3R744A | 5.3 (4.2–6.4) |
1.2 | 6.2 (4.6–9.2) |
1.0 |
| FY 23 | msh3 R744L | NDc | - | 4.1 (3.1–6.1) |
0.6 |
| FY 23 | msh3P745L | ND | - | 5.4 (4.0–8.1) |
0.8 |
| FY 23 | msh3P745A | 4.1 (3.6–5.4) |
0.9 | ND | - |
| FY 23 | msh3R761A | 5.5 (5.1–6.3) |
1.2 | ND | - |
| Walker A residues: | |||||
| FY 23 | msh3G795A | 7.3 (5.6–8.6) |
1.7 | ND | - |
| FY 23 | msh3G796A | 300.5 (226.7–420.3) |
68.7 | 277.0 (174.5–370.7) |
42.6 |
| Aromatic residues: | |||||
| FY 23 | msh3P774A | 5.7 (5.0–7.3) |
1.3 | ND | - |
| FY 23 | msh3Y925A | ND | - | 94.5 (66.1–106.4) |
14.5 |
| FY 23 | msh3F940A | ND | - | 79.98 (62.8–92.8) |
12.3 |
FY23 was co-transformed with msh3 alleles carried on low copy number plasmid (Table S3) and the tetra nucleotide repeat plasmid (pBK1). Mutation rates were determined as described and numbers in parentheses indicate the 95% confidence intervals.
FY23 was co-transformed with msh3 alleles under the control of a galactose-inducible promoter (Table S3) and the tetranucleotide repeat plasmid (pBK1). Cultures were grown for 24 hours in media containing 2% galactose to induce expression of MSH3.
ND = Not Determined
Consistent with this idea, the MMR phenotypes in the presence of the Walker A glycine mutations also tended to be stronger than the 3’ NHTR phenotypes (compare Tables 1 and 2). Mutation of the first glycine to alanine (G795A) resulted in a significant 35-fold increase in mutation rate with the tetranucleotide repeat sequence and a 43-fold increase in the presence of the dinucleotide repeat sequence (Table 2). However, when this residue was changed to aspartic acid, the tetranucleotide repeat mutation rate was only 3-fold higher than in the wild-type background (Table 2). In contrast, when the second, more highly conserved glycine (G796), was changed to either alanine or aspartic acid, there was a dramatic increase in mutation rate (114-fold and 70-fold higher than wild-type, respectively) in the presence of the tetranucleotide repeat sequence (Table 2). msh3G796A also caused a very high mutation rate (166-fold higher than wild-type) in the presence of the dinucleotide repeat sequence (Table 2), consistent with Msh2-Msh3G796A interfering with any Msh2-Msh6-mediated repair of these lesions.
Given the strong phenotype of the G796A mutation in Msh3, we tested this allele for a dominant negative effect on MMR (Table 3) by expressing the allele in a MSH3 background from the low copy plasmid or from an overexpression plasmid. msh3G796A had a strong dominant negative phenotype in MMR, even when expressed in low copy. This is in contrast to the transmitter region alleles and msh3G795A that had no dominant negative effect (Table 3).
In contrast to mutations in the putative nucleotide binding pocket, the MMR phenotype of the transmitter region alleles closely paralleled the phenotypes in 3’ NHTR. msh3R744A showed significant increases in mutation rate relative to wild-type in the presence of tetranucleotide repeat (29-fold) (Table 2, Fig. 3b) and the dinucleotide repeat reporter (16.7-fold) (Table 2), analogous to the defective phenotype in 3’ NHTR (Table 1). When this position was changed instead to leucine (msh3R744L), this led to a null phenotype (~60-fold increase) in tetranucleotide repeat slippage repair (Table 2, Fig. 3b), again, like the null 3’ NHTR phenotype (Table 1). With msh3P745A, we observed a modest four-fold increase in mutation rate (Table 2, Fig. 3c), comparable to the mild defect in 3’ MHTR (Table 1). However, a change at this position to leucine (msh3P745L), an HNPCC mutation found in the analogous position in hMsh2, resulted in a null MMR phenotype in vivo (Table 2), as well as in 3’ NHTR (Table 1). Therefore R744 and P745 of the putative transmitter region of Msh3 are functionally important for both 3’ NHTR and MMR.
As in 3’ NHTR, changes at R761 or N762 resulted in mild phenotypes in MMR. We observed a modest 4-fold increase in mutation rate with msh3R761A, msh3N762A and msh3N762L (Table 2). These data indicated that R761 and N762 contribute to accurate MMR, but neither is essential for function.
FLY motif alleles are proficient in heteroduplex rejection
The different phenotypes of msh3Y925A, msh3F940A and msh3Y942A in MMR and 3’ NHTR indicated different requirements for the regulation of nucleotide binding in the two pathways. This could be a result of different DNA substrates (loop versus flap) and/or the different time scales of each type of repair; MMR, which must coordinate with the replication fork,60 is rapid while 3’ NHTR and DSBR take longer.61 To distinguish between these possibilities, we tested a subset of the msh3 alleles, including the Y and FLY consensus sequences, in preventing homeologous recombination, or heteroduplex rejection (Fig. 1).43 We used an intron-based intramolecular recombination assay that uses a HIS3 reporter.62 In this assay, loss of MSH3 allows recombination to occur between homeologous sequences. The substrate for heteroduplex rejection that we used is predicted to be a 4 base loop (cβ2/cβ2-4L62), as in the MMR slippage assay, but the mechanism occurs over a longer time scale,45–46 more similar to that of 3’NHTR.56
The low copy number MSH3 plasmid complemented the msh3Δ (Table 4), leading to reduced recombination, as previously described.27; 62 In the presence of msh3R744L, a transmitter allele that was defective in both MMR and 3’ NHTR, we observed a null phenotype in heteroduplex rejection, with a 12-fold increase in the rate of homeologous recombination over that observed with MSH3 (Table 4). In contrast, msh3G795D, which showed a very mild MMR defect and a somewhat stronger defect in 3’ NHTR, exhibited wild-type activity in heteroduplex rejection. Notably, msh3Y925A and msh3Y942A, which had null phenotypes in MMR but only mild 3’ NHTR phenotypes, exhibited wild-type or near wild-type level of homeologous recombination in the heteroduplex rejection assay. msh3F940A, which also had a null MMR phenotype, exhibited a stronger defect than msh3Y925A and msh3Y942A in heteroduplex rejection but still retained some activity, similar to its 3’ NHTR phenotype. These results indicated that it was not the DNA substrate that altered the requirement for the FLY motif but instead it may be the kinetics of repair that led to these differences.
Table 4.
Rates of homeologous recombination with msh3 alleles.
| Strain | Plasmid | Rate of homeologous recombinationa(x 10−7) |
Fold Change |
|---|---|---|---|
| EAY1601 (MSH3) | - | 1.42 (1.0–2.2) |
1 |
| EAY1613 (msh2Δ) | - | 69.5 (59.2–86.4) |
49 |
| EAY 1625 (msh3Δ) | - | 47.88 (44.7–51.3) |
33.7 |
| EAY1625 | MSH3b | 5.7 (1.6–10.9) |
1 |
| EAY1625 | Empty Vector | 51.6 (44.5–81.1) |
9.0 |
| Transmitter residues: | |||
| EAY1625 | msh3R744L | 67.5 (47.7–182.3) |
11.8 |
| Walker A residues: | |||
| EAY1625 | msh3G795D | 5.6 (2.5–7.4) |
0.9 |
| Aromatic residues: | |||
| EAY1625 | msh3Y925A | 5.5 (2.1–6.8) |
0.9 |
| EAY1625 | msh3F940A | 33.1 (28.7–36.1) |
5.8 |
| EAY1625 | msh3Y942A | 17.5 (11.0–22.1) |
3.0 |
Rate of homeologous recombination was calculated in strains predicted to form recombination intermediates that contain a four base loop. Numbers in parentheses represent 95% confidence intervals. An increase in the rate of homeologous recombination indicates a decrease in heteroduplex rejection and therefore a decrease in Msh2-Msh3 activity.
EAY1625 was transformed with low copy number plasmids bearing MSH3 or msh3 alleles.
Discussion
In this study, we targeted ten positions within Msh3 that were predicted to be important in regulating the conformational changes that lead to coordinated nucleotide binding, exchange and hydrolysis following mispair or IDL binding in order to complete MMR. In general, residues that were required for 3’ NHTR were also required for MMR, indicating that the structural and molecular requirements are similar in the two pathways. However, mutations in the conserved Y and FLY motifs had only mild effects on 3’ NHTR but were completely deficient in Msh2-Msh3-mediated MMR. These same mutations were similarly functional in heteroduplex rejection, allowing us to distinguish between possible explanations for the differences in requirements for this region of Msh3 (Fig. 5).
Figure 5. Summary of msh3 allele phenotypes in the in vivo pathways examined in this study.
Each allele was characterized as having wild-type function (++; > 90% of MSH3 activity), intermediate phenotype (+; between 20 and 90% of MSH3 activity) or a defective phenotype (−; < 20% MSH3 activity).
Communication between the mispair binding and ATPase domains
The transmitter region is at the interface of the lever domain (Domains II and III; Fig. 2) and the ATPase domain (Domain V; Fig. 2) and has been implicated in facilitating communication between the two ends of Msh complexes.10; 30; 32; 59; 63 It has been noted that several residues at the interfaces of the lever and DNA binding domains and the lever and ATPase domains adopt different conformations when T. aquaticus MutS is bound to mismatch DNA compared to when it is unbound,32 including the ATP binding pocket made up of the FLY and YUP motifs (Fig 3a and data not shown). These observations are consistent with the induction of conformational changes upon DNA binding that influence events at the ATPase domain. Importantly, of the ten HNPCC mutations present in the lever domain of hMSH2, seven trace to the transmitter region,59 which is the region we have focused on in this study. Three of the four positions that we tested in the putative Msh3 transmitter region (P745, R761 and N762) were mutated at the analogous positions in hMSH2 in HNPCC patients. Our mutational analyses indicate that the R744 and P745 residues are required for both MMR and 3’ NHTR (and heteroduplex rejection) indicating that the communication of substrate binding occurs in a similar manner in these diverse pathways (Fig. 5). These mutations may result in faulty communication, leading to the inability of Msh2-Msh3 to proceed efficiently to the next step in repair, including appropriate nucleotide binding. Mutational and biochemical analyses demonstrated that P622L of hMsh2 (analogous to P745L in yMsh3) results in defective DNA-binding and ATPase functions,64 but msh2P622L in human cell culture and in yeast is destabilized, which may also account for an HNPCC phenotype.65–66 Western blots indicated that msh3P745L is stably expressed in yeast (Fig. S1) therefore this does not account for the null phenotype in Msh2-Msh3 function (Table 1).
We modeled the changes at these positions, based on the hMSH2-hMSH3 crystal structure.30 The structure predicted that Msh3 R744 makes polar contacts with E471 and D475 residues in the lever domain (Fig. 3c, left panel) and modeling predicted that these contacts will be disrupted by a change to alanine (Fig. 3c, middle panel) or to leucine (Fig. 3c, right panel), which may explain the defects in repair observed with these alleles (Tables 1 and 2). In contrast, modeling indicated that P745 made no polar contacts and when changed to alanine, there was no notable difference in that region (Fig. 3c), consistent with largely proficient repair (Tables 1 and 2). However a change to leucine at this position is predicted to gain a polar contact with V817 in the ATPase domain (Fig. 3c), perhaps explaining the 3’ NHTR and MMR defects with this allele (Tables 1 and 2; Fig. 5).
Interestingly, mutations at two of the positions that align with positions in hMsh2 that are implicated in HNPCC (yMsh3 R761 and N762; Fig. 2a) did not exhibit a strong mutator phenotype or a defect in 3’ NHTR, consistent with the wild-type phenotype of msh2H658Y in S. cerevisiae (equivalent to msh3N762)67. The structural model of the R761 microenvironment indicated that main chain contacts with E766 and P818 are not disrupted by any amino acid substitution (data not shown). Similarly, N762 makes polar contacts with I764 and E766 that did not appear to be affected by these changes. It is nonetheless possible that these residues help modulate communication. Over time a defect could manifest a significant mutator phenotype, as has been suggested for the deletion of the N-terminus of Msh2.68–69 Of note is the observation that a S656P change in yMsh2 (analogous to S760 in yMsh3, adjacent to the residues we tested) leads to defective Msh2-Msh6-mediated MMR and a dominant negative phenotype, indicating that it blocks the function of wild-type Msh2-Msh6.11 Notably, msh2S656P was largely unaffected in 3’ NHTR (an Msh2-Msh3-specific activity) similar to our observations with mutations at Msh3 R761 and N762. Therefore these residues may be less important for Msh2-Msh3 function than for Msh2-Msh6 function.
Walker A glycines are critical for Msh2-Msh3 function
The Walker A motif , or P-loop, in proteins with ATPase activity is thought to bind the phosphate moieties of ATP, a prerequisite for catalysis.70 The Walker A motif in Msh proteins is GXXXG/AGKS/T (Fig 2a) and the GKS motif in Msh2 was demonstrated to be critical for Msh2-Msh6 and Msh2-Msh3 functions.11; 71 It was shown to be similarly important in MutS and Msh6.72–74 In Msh3 the Walker A motif includes a glycine pair preceding the conserved lysine (GXXXGGKS). Both glycines are important for Msh2-Msh3 function in 3’ NHTR and MMR, but to differing extents (Tables 1 and 2; Fig. 5). However, in both pathways, the alanine change led to a stronger defect than the change to aspartic acid. Interestingly, this position of T. aquaticus MutS and yMsh6 is occupied by an alanine. One possibility is that the regulation of Msh3 ATP binding and/or hydrolysis in msh3G795A is more like that of Msh6, altering the coordination of events within Msh2-Msh3; regulation of nucleotide binding and hydrolysis appears to be distinct in Msh2-Msh6 and Msh2-Msh3.24–25
As expected, msh3G796D exhibited a null phenotype in 3’ NHTR (Table 1). Intriguingly, while deficient in 3’ NHTR, msh3G796A did not exhibit a complete null phenotype in DSBR. In contrast, both msh3G796A and msh3G796D had very high mutation rates (Table 2) and msh3G796A displayed a strong dominant negative phenotype (Table 3). Therefore the requirements for ATP binding appear less stringent in 3’ NHTR. We predict that alteration of G796 allows binding of mispairs, but blocks ATP binding and hydrolysis, thereby resulting in complexes that do not form a sliding clamp or turnover and remain “stuck” at the mispair, explaining the dominant negative phenotype. Consistent with this, the analogous yMsh2 mutation in (G693D) allowed binding of msh2G693D-Msh6 and msh2G693D-Msh3 to specific substrates,13; 26 but msh2G693D-Msh6 was inhibited for ATP binding13 and msh2G693D-Msh3 inhibited dissociation of the mutant complex from specific DNA substrates.26
Regulation and coordination of nucleotide binding
Once information about mispair or 3’ flap binding by Msh2-Msh3 has been transmitted, conformational changes in the nucleotide binding pocket are thought to regulate nucleotide exchange and/or hydrolysis within both Msh2 and Msh3; proper regulation of these events is critical for function.24–25 P774, within the YUP motif, resides on a loop opposite the FLY motif loop and was predicted to influence the conformation of the nucleotide binding site.30 Surprisingly, we observed no defect in 3’ NHTR or MMR when this highly conserved proline residue was changed to alanine or leucine (Tables 1 and 2; Fig. 5), suggesting that this position is not stringent in its amino acid requirement, although we have not made a comprehensive suite of mutations at this position. Structural modeling indicated no changes in microenvironment when P774 was changed to alanine or leucine (Fig 4c).
Based on the crystal structures of hMSH2-hMSH3,30 hMSH2-hMSH659 and MutS32; 63, the aromatic-rich YUP and FLY motifs (Fig. 2a) were found to be closely associated in three-dimensional space to form a nucleotide binding pocket. A nucleotide sandwich between the Y of the YUP motif (Y772 in yMsh3) and the Y of the FLY motif (Y942 in yMsh3) was proposed, with the F of the FLY motif (F940 in yMsh3) locking in the bound nucleotide.30 In the Msh3 nucleotide-free state in the crystal structure, a unique Msh3-specific aromatic residue (Y925 in yMsh3) pushes the F of the FLY motif into the ATP binding site, thereby blocking nucleotide-binding and stabilizing the nucleotide-free state.30 It is unclear how Msh3 would be able to adopt an ATP-bound state upon DNA binding. One possibility is that a conformational change, perhaps within one of the transmitter regions, forces the phenylalanine out of the binding site, making the pocket more accessible to nucleotide. Consistent with the FLY motif playing a critical role in Msh2-Msh3 function, when either the F (F940) or the Y (Y942) was changed to alanine MMR was completely abolished (Table 2). Similarly, mutating Y925 to alanine, removing that aromatic group that is unique to Msh3, resulted in a null MMR phenotype (Table 2). Furthermore, msh3Y925A and msh3F940A exhibited a dominant negative phenotype (Table 3), indicating that these mutant complexes block the activity of wild-type complex.
Previous work30 and our own structural modeling predicted that these alleles would make the nucleotide binding pocket more accessible. Changing either Y925 or F940 to much smaller residue like alanine was predicted to prevent occlusion of the nucleotide binding pocket (compare Fig. 4a, d and e) and the Y942A mutation was predicted to remove one side of the nucleotide sandwich (compare Fig. 4a and f). Such changes could alter nucleotide occupancy and override the wild-type sequence of conformational changes mediated in part by nucleotide (ADP or ATP) binding that coordinates proper MMR. A more accessible Msh3 nucleotide-binding pocket in the absence of these aromatic residues, might allow premature ATP-binding by Msh3 or lead to the formation of a non-productive Msh3-ADP bound complex.24
Differential molecular requirements of Msh2-Msh3 in 3’NHTR and heteroduplex rejection
While msh3Y925A, msh3F940A and msh3Y942A had severe MMR defects (Table 2), the effect on 3’ NHTR was quite mild (Table 1). It is intriguing that the requirements for residues within the FLY region are distinct in MMR and 3’NHTR, while the transmitter region and the Walker A motif requirements are more similar. This suggests that the coordination of nucleotide binding and exchange within the complex is not as critical in 3’ NHTR as in MMR. One possible explanation for this is the difference in the kinetics of MMR versus 3’ NHTR. At the replication fork, MMR has to occur in a matter of seconds, prior to the re-establishment of chromatin structure.75 In contrast, 3’ NHTR occurs more slowly. Southern blot analyses indicated that repair occurs over the course of several hours at the wild-type MAT locus.56; 61; 76 We tested this possibility by examining the effect of the msh3Y925A and FLY alleles on heteroduplex rejection, a Msh2-Msh3-specific pathway that detects the same 4 loop substrate as in the slippage assays,62 but has a time-scale similar to that of 3’ NHTR.45–46 The wild-type phenotypes of msh3Y925A, msh3F940A and msh3Y942A in heteroduplex rejection (similar to 3’ NHTR) indicated that the substrate (loop versus flap) did not lead to distinct conformational changes that obviated the need for regulation of the nucleotide binding pocket. Furthermore, while nucleotide binding is essential for interactions between MutS and MutL homologs,23 it is unlikely that the interaction with the downstream MMR-specific Mlh1-Pms1 complex is the sole factor in the differences in phenotypes between MMR and 3’ NHTR; heteroduplex rejection also involves MLH1, although not to the same extent as MSH2, MSH3 or MSH6.62 Instead, the requirement for ATP binding itself appears to be different in MMR versus 3’ NHTR and heteroduplex rejection (Fig. 5). While ATP binding by Msh3 was required for both MMR and 3’ NHTR (see msh3G796D phenotypes), the FLY motif mutation phenotypes suggested that the conformation of the binding pocket and therefore of the bound ATP is critical for MMR. In contrast, more flexible ATP binding, i.e. ATP bound in different positions or conformations, may be acceptable in 3’ NHTR. There may simply be more time for the ATP to stochastically find the “right” position for function in 3’ NHTR (or heteroduplex rejection). Alternatively, the sequence of conformational changes within Msh2-Msh3 may be distinct in 3’ NHTR, because of different requirements for ATP hydrolysis and/or protein-protein interactions, making the precise positioning and/or timely binding of ATP less important.
It is notable that the 3’NHTR phenotypes for msh3Y925 and msh3Y942 are very mild, while msh3F940 exhibited a stronger phenotype. If Y925 is in fact pushing F940 into the ATP-binding site to stabilize a nucleotide-free state, this does not appear to be critical for 3’ NHTR, while other functions of F940 are more important. These distinct requirements will be the focus of future investigations.
Materials and Methods
Plasmids and yeast strain construction
All yeast transformations were performed using the lithium acetate method.77 All strains are in the S288c background (FY2378) and are listed in supplementary tables.
The low copy number MSH3 plasmid was made in pEAA378, a derivative of pRS414 that carries the NATMX cassette in place of TRP1. MSH3 and 1kb upstream and 0.3 kb downstream of start and stop sites, was PCR amplified from pEAI215.27 The PCR primers introduced BamHI and SacII sites and the resulting fragment was digested with these enzymes and ligated into pEAA378 to generate pGW2. The PCR fragment was confirmed by DNA sequencing. The msh3 alleles were made by PCR-based site-directed mutagenesis (oligonucleotides used are listed in Table S2), confirmed by DNA sequencing and sub-cloned into pGW2. For galactose inducible overexpression of msh3 alleles, a Bsu36I-MluI fragment containing each allele was sub-cloned into pMMR2079, behind the GAL10 promoter. The plasmids carrying msh3 alleles are listed in Table S3.
All structural modeling was performed using PyMOL (The Pymol molecular Graphics system, Schrödinger, LLC.80
Double strand break survival and mating type switching assay
EAY1118 encodes a double non-homology at the MAT locus and carries a galactose-inducible HO endonuclease that creates a DSB at the MAT locus.53 This strain was transformed with low copy plasmids expressing msh3 alleles and the efficiency of DSBR was assayed as described previously.56 All strains used in this assay are listed in Table S5. Briefly, cultures were grown to mid-log phase in the presence of lactate as carbon source. DSBs were induced by the addition of galactose for 45 minutes. Cultures were washed, diluted and plated onto YPD (1% yeast extract, 2% bacto-peptone, and 2% dextrose) + clonNAT (Werner; 100 µg/ml) plates. After incubation at 30°C for three days, percent survival was calculated as the ratio of number of colonies that grew following DSB induction relative to uninduced control. At least three independent transformants were tested for each msh3 allele and each set was tested in duplicate. To determine mating type switching, 10–30 individual colonies that survived DSB induction, from each transformant, were mated with FY23 (MATa) and FY86 (MATα) on YPD and replica plated on synthetic minimal media lacking lysine and leucine to select for diploids. Those cells that were able to mate with FY23 (MATa) had switched from MATa to MATα following induction of the DSB and were therefore competent for DSBR.
Determination of mutation rates
EAY420 (msh3Δ, see Table S1) was co-transformed with pSH44((GT)16.5 repeat) or pBK1 ((CAGT)16 repeat)4 and a low copy number plasmid carrying MSH3 or the msh3 alleles. Tranformants were selected on synthetic deficient(SD) media plates lacking tryptophan and containing clonNAT (100µg/ml) (SD-trp+NAT). In these plasmids a dinucleotide or tetranucleotide repeat sequence ((GT)16.5 or (CAGT)16, respectively) was inserted in-frame upstream of URA3. In the presence of a DNA slippage event that is not repaired, resulting in an IDL, the URA3 gene is put out of frame; these events can be selected in the presence of 5 -FOA. All strains used for determination of mutation rates are listed in Table S4.
Slippage assays were performed as described previously.4; 27 Briefly, saturated overnights were grown up from at least 7 individual colonies, from each of three independent transformants of each msh3 allele. Appropriate dilutions were plated on selective plates lacking tryptophan and containing clonNAT (100 µg/ml) and 5FOA (µg/ml) (−trp+NAT+5FOA) and permissive (−trp+NAT) plates. Plates were incubated at 30°C for three days and colonies were counted. Mutation rates were calculated by the method of the median.81 95% confidence intervals were from tables calculated by Nair (1940)82 and Dixon and Massey (1969).83
The mutation rates of msh3Δ carrying the MSH3 plasmid or the empty vector were lower than in the wild-type MSH3 or the msh3Δ background in the absence of plasmid (Table 2). This is likely a result of the slower growth rate of the strain in the presence of clonNAT that was used to select for the presence of the msh3 plasmids. The doubling time of MSH3 or msh3Δ strains carrying any of the NATMX plasmids decreased by about 1.5-fold (data not shown). These strains will therefore have gone through fewer generations in the same, defined time period than strains without plasmid, leading to an apparently lower mutation rate. Nonetheless, the fold change in mutation rate between wild-type and msh3Δ is essentially the same.
The mutation rates in Table 1 were consistently lower than in Table 2 and the range between MSH3 and msh3Δ is also reduced. We attribute this to the different strain backgrounds used in each data set. Consistent with this possibility, the EAY1118 background grows significantly more slowly than EAY420, with or without the NATMX plasmids.
For determination of dominant negative phenotype, wild-type strain (FY23; MSH3) was co-transformed with pBK1 and plasmids carrying the msh3 alleles under the control of the galactose-inducible promoter. Strains used are listed in Table S7. Saturated cultures were grown up in synthetic minimal media containing galactose as carbon source to induce the expression of msh3 alleles. Assays were performed as described above.
Homeologous recombination assays
EAY162527 (msh3Δ, see Table S1) encodes substrates that are predicted to form intermediates containing 4 nucleotide loops, substrates for homeologous recombination, or heteroduplex rejection. This strain was transformed with a low copy plasmid encoding MSH3 or msh3 alleles. All strains used to measure homeologous recombination are listed in Table S8. 14–21 single colonies from each strain grown on YPD +clonNAT plates were inoculated into 5 ml of YPGG (1% yeast extract, 2% bacto-peptone, 4% galactose, and 2% glycerol) medium in the presence of clonNAT and grown for 50 hours at 30 °C. Appropriate dilutions of cells were plated onto synthetic galactose medium lacking histidine (selective) and onto synthetic complete medium (permissive), both with clonNAT to maintain the msh3 plasmid. Plates were incubated for 4 days at 30 °C and then scored for frequency of His+ colonies. The rate of homeologous recombination was calculated as described.62
Supplementary Material
Highlights.
Msh2-Msh3 participates in the removal of 3’ non-homologous tails in recombination.
Msh2-Msh3 initiates mismatch repair (MMR) of DNA slippage events in replication.
Msh2-Msh3 undergoes conformational changes in ATP binding pocket upon DNA binding.
Mutations in the Msh3 ATP binding pocket disrupt MMR but not recombination.
Kinetic differences in the pathways may dictate distinct ATP positioning constraints.
Acknowledgments
We thank Dr. Wei Yang for helpful discussion and Dr. Mark Sutton for discussions and for reviewing the manuscript prior to submission. We thank members of the Surtees lab for technical assistance, especially Dr. Andrew Bukata for statistical analyses and Bangchen Wang for cloning, and for critical discussions. We thank Dr. Justin Hetzel for help with the modeling. Work in the Surtees lab is supported by NIH GM087459.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mechanisms of Ageing and Development. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Dong J, Pan X, Oum J-H, Boeke JD, Lee SE. Microarray-Based Genetic Screen Defines SAW1, a Gene Required for Rad1/Rad10-Dependent Processing of Recombination Intermediates. Molecular Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sia E, Kokoska R, Dominska M, Greenwell P, Petes T. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen LE, Jauert PA, Kirkpatrick DT. The Large Loop Repair and Mismatch Repair Pathways of Saccharomyces cerevisiae Act on Distinct Substrates During Meiosis. Genetics. 2005;170:1033–1043. doi: 10.1534/genetics.104.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 Acts in Repair of Base-Base Mispairs. Mol. Cell. Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinen CD. Genotype to phenotype: Analyzing the effects of inherited mutations in colorectal cancer families. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2010;693:32–45. doi: 10.1016/j.mrfmmm.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hargreaves VV, Shell SS, Mazur DJ, Hess MT, Kolodner RD. Interaction between the Msh2 and Msh6 Nucleotide-binding Sites in the Saccharomyces cerevisiae Msh2-Msh6 Complex. Journal of Biological Chemistry. 2010;285:9301–9310. doi: 10.1074/jbc.M109.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hargreaves VV, Putnam CD, Kolodner RD. Engineered Disulfide-forming Amino Acid Substitutions Interfere with a Conformational Change in the Mismatch Recognition Complex Msh2-Msh6 Required for Mismatch Repair. Journal of Biological Chemistry. 2012;287:41232–41244. doi: 10.1074/jbc.M112.402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junop MS, Oblomova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 11.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase Activities Are Both Required during Mismatch Repair. Mol. Cell. Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studamire B, Price G, Sugawara N, Haber JE, Alani E. Separation-of-Function Mutations in Saccharomyces cerevisiae MSH2 That Confer Mismatch Repair Defects but Do Not Affect Nonhomologous-Tail Removal during Recombination. Mol. Cell. Biol. 1999;19:7558–7567. doi: 10.1128/mcb.19.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kijas AW, Studamire B, Alani E. msh2 Separation of Function Mutations Confer Defects in the Initiation Steps of Mismatch Repair. Journal of Molecular Biology. 2003;331:123–138. doi: 10.1016/s0022-2836(03)00694-6. [DOI] [PubMed] [Google Scholar]
- 14.Alani E, Lee JY, Schofield MJ, Kijas AW, Hsieh P, Yang W. Crystal Structure and Biochemical Analysis of the MutS{middle dot}ADP{middle dot}Beryllium Fluoride Complex Suggests a Conserved Mechanism for ATP Interactions in Mismatch Repair. J. Biol. Chem. 2003;278:16088–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]
- 15.Gradia S, Subramanian, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp in mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 16.Gradia S, Acharya S, Fishel R. The Human Mismatch Recognition Complex hMSH2-hMSH6 Functions as a Novel Molecular Switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 17.Gradia S, Acharya S, Fishel R. The Role of Mismatched Nucleotides in Activating the hMSH2-hMSH6 Molecular Switch. J. Biol. Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- 18.Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL, Kolodner RD. Probing DNA- and ATP-mediated Conformational Changes in the MutS Family of Mispair Recognition Proteins Using Deuterium Exchange Mass Spectrometry. Journal of Biological Chemistry. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess MT, Das Gupta R, Kolodner RD. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J. Biol. Chem. 2002;277:25545–25553. doi: 10.1074/jbc.M202282200. [DOI] [PubMed] [Google Scholar]
- 20.Hess MT, Mendillo ML, Mazur DJ, Kolodner RD. Biochemical basis for dominant mutations in the Saccharomyces cerevisiae MSH6 gene. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:558–563. doi: 10.1073/pnas.0510078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinen CD, Cyr JL, Cook C, Punja N, Sakato M, Forties RA, Martin Lopez J, Hingorani MM, Fishel R. hMSH2 controls ATP processing by hMSH2-hMSH6. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M111.297523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acharya S, Foster PI, Brooks R, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel TA, Erie DA. DNA MISMATCH REPAIR. Annual Review of Biochemistry. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 24.Owen BAL, H Lang W, McMurray CT. The nucleotide binding dynamics of human Msh2-Msh3 are lesion dependent. Nat Struct Mol Biol. 2009;16:550–557. doi: 10.1038/nsmb.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian L, Gu L, Li G-M. Distinct Nucleotide Binding/Hydrolysis Properties and Molar Ratio of MutS{alpha} and MutS{beta} Determine Their Differential Mismatch Binding Activities. J. Biol. Chem. 2009;284:11557–11562. doi: 10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surtees JA, Alani E. Mismatch Repair Factor Msh2-Msh3 Binds and Alters the Conformation of Branched DNA Structures Predicted to form During Genetic Recombination. Journal of Molecular Biology. 2006;360:523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 27.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae Msh2-Msh3 and MSH2-MSH6 Complexes Display Distinct Requirements for DNA Binding Domain I in Mismatch Recognition. Journal of Molecular Biology. 2007;366:53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proceedings of the National Academy of Sciences. 2007;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowen JM, Putnam CD, Kolodner RD. Functional Studies and Homology Modeling of Msh2-Msh3 Predict that Mispair Recognition Involves DNA Bending and Strand Separation. Molecular and Cellular Biology. 2010;30:3321–3328. doi: 10.1128/MCB.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat Struct Mol Biol. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, Weninger KR. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 2012;31:2528–2540. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 33.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the Interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 Complexes with DNA Using a Reversible DNA End-blocking System. J. Biol. Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of High-Affinity and Sliding Clamp Modes for MSH2-MSH6 by Single-Molecule Unzipping Force Analysis. Molecular Cell. 2005;20:771–781. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li G-M. Reconstitution of 5'-Directed Human Mismatch Repair in a Purified System. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 37.Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of Msh6 ATPase Activity by Mispaired DNA Induces a Msh2(ATP)-Msh6(ATP) State Capable of Hydrolysis-Independent Movement along DNA. Molecular Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Owen BAL, Yang Z, Lai M, Gajec M, Badger Jd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 39.Wilson T, Guerrette S, Fishel R. Dissociation of Mismatch Recognition and ATPase Activity by hMSH2-hMSH3. J. Biol. Chem. 1999;274:21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 40.van den Broek WJAA, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJTA, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch–repair proteins. Human Molecular Genetics. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 41.Foiry L, Dong L, Savouet C, Hubert L, Te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genetics. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 42.Kantartzis A, Williams Gregory M, Balakrishnan L, Roberts Rick L, Surtees Jennifer A, Bambara Robert A. Msh2-Msh3 Interferes with Okazaki Fragment Processing to Promote Trinucleotide Repeat Expansions. Cell Reports. 2012;2:216–222. doi: 10.1016/j.celrep.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: key reglators of genetic recombination. Cytogenetic and Genome Research. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 44.Evans E, Alani E. Roles for Mismatch Repair Factors in Regulating Genetic Recombination. Mol. Cell. Biol. 2000;20:7839–7844. doi: 10.1128/mcb.20.21.7839-7844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proceedings of the National Academy of Sciences. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldfarb T, Alani E. Distinct Roles for the Saccharomyces cerevisiae Mismatch Repair Proteins in Heteroduplex Rejection, Mismatch Repair and Nonhomologous Tail Removal. Genetics. 2005;169:563–574. doi: 10.1534/genetics.104.035204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirkpatrick DT, Petes TD. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 48.Kearney HM, Kirkpatrick DT, Gerton JL, Petes TD. Meiotic Recombination Involving Heterozygous Large Insertions in Saccharomyces cerevisiae: Formation and Repair of Large, Unpaired DNA Loops. Genetics. 2001;158:1457–1476. doi: 10.1093/genetics/158.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bardwell AJ, Bardwell L, Johnson DK, Friedberg EC. Yeast DNA recombination and repair proteins Rad1 and Rad10 consititute a complex in vivo mediated by localized hydrophobic domains. Mol. MicroBiol. 1993;8:1177–1188. doi: 10.1111/j.1365-2958.1993.tb01662.x. [DOI] [PubMed] [Google Scholar]
- 50.Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 51.Manna AC, Pai KS, Bussiere DE, Davies C, White SW, Bastia D. Helicase-contrahelicase interaction and the mechanism of termination of DNA replication. Cell. 1996;87:881–891. doi: 10.1016/s0092-8674(00)81995-9. [DOI] [PubMed] [Google Scholar]
- 52.Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proceedings of the National Academy of Sciences. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyndaker AM, Alani E. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. BioEssays. 2009;31:315–321. doi: 10.1002/bies.200800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paques F, Haber JE. Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans E, Sugawara N, Haber JE, Alani E. The Saccharomyces cerevisiae Msh2 Mismatch Repair Protein Localizes to Recombination Intermediates In Vivo. Molecular Cell. 2000;5:789–799. doi: 10.1016/s1097-2765(00)80319-6. [DOI] [PubMed] [Google Scholar]
- 56.Lyndaker AM, Goldfarb T, Alani E. Mutants Defective in Rad1-Rad10-Slx4 Exhibit a Unique Pattern of Viability During Mating-Type Switching in Saccharomyces cerevisiae. Genetics. 2008;179:1807–1821. doi: 10.1534/genetics.108.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li F, Dong J, Eichmiller R, Cory H, Minca E, Prakash R, Sung P, Yong SE, Surtees JA, Lee SE. Role of Saw1 in Rad1/Rad10 Complex Assembly at Recombination Intermediates in Budding Yeast. EMBO J. In Press doi: 10.1038/emboj.2012.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 59.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese Lorena S. Structure of the Human MutS[alpha] DNA Lesion Recognition Complex. Molecular Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 60.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch Repair, But Not Heteroduplex Rejection, Is Temporally Coupled to DNA Replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldfarb T, Alani E. Chromatin immunoprecipitation to investigate protein-DNA interactions during genetic recombination. Methods Mol. Biol. 2004;262:223–237. doi: 10.1385/1-59259-761-0:223. [DOI] [PubMed] [Google Scholar]
- 62.Nicholson A, Hendrix M, Jinks-Robertson S, Crouse GF. Regulation of Mitotic Homeologous Recombination in Yeast: Functions of Mismatch Repair and Nucleotide Excision Repair Genes. Genetics. 2000;154:133–146. doi: 10.1093/genetics/154.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HHK, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G[middot]T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 64.Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–478. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 65.Mastrocola AS, Heinen CD. Lynch syndrome-associated mutations in MSH2 alter DNA repair and checkpoint response functions in vivo. Human Mutation. 2010;31:E1699–E1708. doi: 10.1002/humu.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gammie AE, Erdeniz N, Beaver J, Devlin B, Nanji A, Rose MD. Functional Characterization of Pathogenic Human MSH2 Missense Mutations in Saccharomyces cerevisiae. Genetics. 2007;177:707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polaczek P, Putzke A, Leong K, GA B. Functional genetic tests of DNA mismatch repair protein activity in Saccharomyces cerevisiae. Gene. 1998;213:159–167. doi: 10.1016/s0378-1119(98)00150-4. [DOI] [PubMed] [Google Scholar]
- 68.Kumar C, Piacente SC, Sibert J, Bukata AR, O'Connor J, Alani E, Surtees JA. Multiple Factors Insulate Msh2-Msh6 Mismatch Repair Activity from Defects in Msh2 Domain I. Journal of Molecular Biology. 2011;411:765–780. doi: 10.1016/j.jmb.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cyr JL, Brown GD, Stroop J, Heinen CD. The predicted truncation from a cancer-associated variant of the MSH2 initiation codon alters activity of the MSH2-MSH6 mismatch repair complex. Molecular Carcinogenesis. 2012;51:647–658. doi: 10.1002/mc.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanson L, May L, Tuma P, Keeven J, Mehl P, Ferenz M, Ambudkar SV, Golin J. The Role of Hydrogen Bond Acceptor Groups in the Interaction of Substrates with Pdr5p, a Major Yeast Drug Transporter†. Biochemistry. 2005;44:9703–9713. doi: 10.1021/bi0502994. [DOI] [PubMed] [Google Scholar]
- 71.Drotschmann K, Clark AB, Kunkel TA. Mutator phenotypes of common polymorphisms and missense mutations in MSH2. Current Biology. 1999;9:907–910. doi: 10.1016/s0960-9822(99)80396-0. [DOI] [PubMed] [Google Scholar]
- 72.Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding functions. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu TH, Marinus MG. Dominant negative mutator mutations in the mutS gene of Escherichia coli. Journal of Bacteriology. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSa. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conde F, Refolio E, CordÃ3n-Preciado V, Cortés-Ledesma F, Aragón L, Aguilera As, San-Segundo PA. The Dot1 Histone Methyltransferase and the Rad9 Checkpoint Adaptor Contribute to Cohesin-Dependent Double-Strand Break Repair by Sister Chromatid Recombination in Saccharomyces cerevisiae. Genetics. 2009;182:437–446. doi: 10.1534/genetics.109.101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holmes A, Haber J. Physical Monitoring of HO-Induced Homologous Recombination. In: Henderson D, editor. DNA Repair Protocols. Vol. 113. Humana Press; 1999. pp. 403–415. [DOI] [PubMed] [Google Scholar]
- 77.Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Research. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winston F, Dollard C, SL R-H. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 79.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Current Biology. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 80.Delano W. The PyMOL User's Manual. San Carlos, CA USA: Delano Scientific; 2002. [Google Scholar]
- 81.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nair KR. Table of Confidence Interval for the Median in Samples from Any Continuous Population. SankhyÄ• : The Indian Journal of Statistics (1933–1960) 1940;4:551–558. [Google Scholar]
- 83.Dixon W, Massey F. Introduction to Statistical Analysis. McGraw Hill, New York: 1969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.