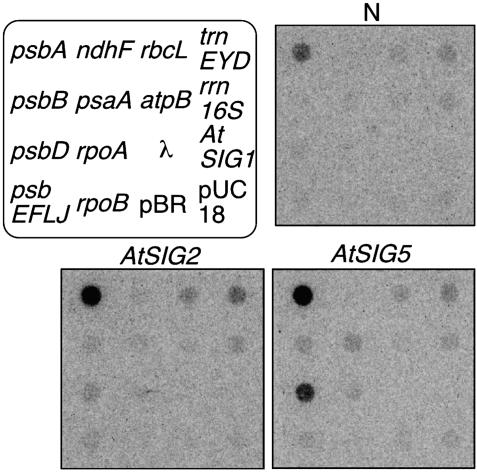
Fig. 3.
Autoradiograms representing run-on transcription activities of plastid-encoded genes in dark-adapted protoplasts to overexpress AtSig2 or AtSig5. α-32P-labeled run-on transcripts derived from 25 μl of protoplast suspension (a density of 2.5 mg chlorophyll per ml) were hybridized to dot blots containing DNA fragments specific for plastid-encoded genes including psaA, psbA, psbD, psbE-F-L-J, ndhF, rbcL, atpB, rpoA, rpoB, trnE-Y-D, and rrn16S genes that encode P700 apoprotein A1 of PSI, D1, D2, CP47, small subunit proteins of PSII, NADH dehydrogenase NDS, the large subunit of riblose-1,5-bisphosphate carboxylase/oxygenase, α-subunit of ATPase, α,β-subunit of PEP, the set of transfer RNAs, and 16S ribosomal RNA, respectively. To detect transformed vector sequences, pUC18 vector DNA (pUC) was used as a control. MspI-digested pBR322 DNA fragments (pBR) and lambda DNA (λ) probes indicated that there were no nonspecific transcripts. Because no transcription signal was detected from AtSig1 probe, nuclear RNA polymerase is unlikely involved in the protoplast run-on transcription activity.