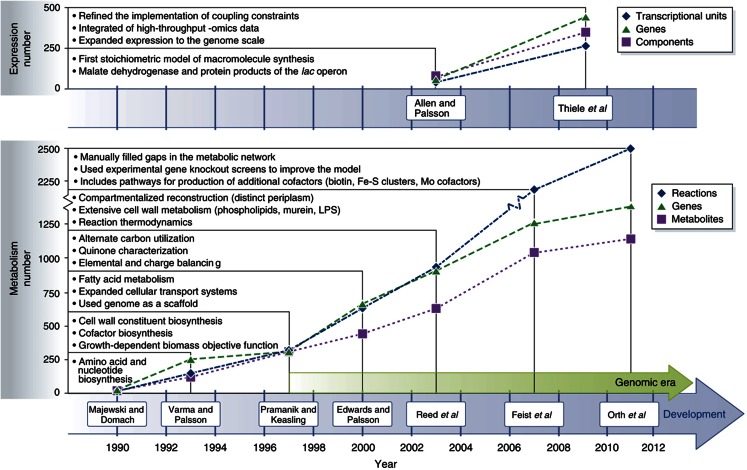
Figure 1.
History of the E. coli expression and metabolic reconstructions. Shown in the upper portion of the graph are 2 milestone efforts contributing to the reconstruction of the E. coli transcription and translation network, and shown in the bottom portion of the graph are 7 milestone efforts contributing to the reconstruction of the E. coli metabolic network. For each of the two reconstructions shown (Allen and Palsson, 2003; Thiele et al, 2009) in the upper graph, the number of included transcription units (blue diamonds), genes (green triangles) and components (purple squares) are displayed. For each of the seven reconstructions shown (Majewski and Domach, 1990; Varma and Palsson, 1993; Pramanik and Keasling, 1997; Edwards and Palsson, 2000; Reed et al, 2003; Feist et al, 2007; Orth et al, 2011) in the bottom graph, the number of included reactions (blue diamonds), genes (green triangles) and metabolites (purple squares) are displayed. Moreover, listed is noteworthy content expansion that each successive reconstruction provided over previous efforts. For example, Varma et al (1993), and Varma and Palsson (1993) included amino acid and nucleotide biosynthesis pathways in addition to the content that Majewski and Domach (1990) characterized. The start of the genomic era (Blattner et al, 1997) marked a significant increase in included components for successive iterations of the network reconstruction. The significant increase in the number of reactions in 2007 (Feist et al, 2007) was, in large part, due to the removal of many lumped reactions, which were often included for lipid and cell wall biosynthesis in earlier metabolic reconstructions. Thiele et al (2009) expanded the initial work of Allen and Palsson (2003) by increasing the scope of the transcription and translation network from a few example pathways to all known genes involved in protein synthesis (i.e., expression). Not included on the timeline is a metabolic reconstruction based upon Reed et al (2003), which was modified to include additional reactions from the KEGG (Kanehisa et al, 2008) database and incorporated into the MetaFluxNet software package (Lee et al, 2005).