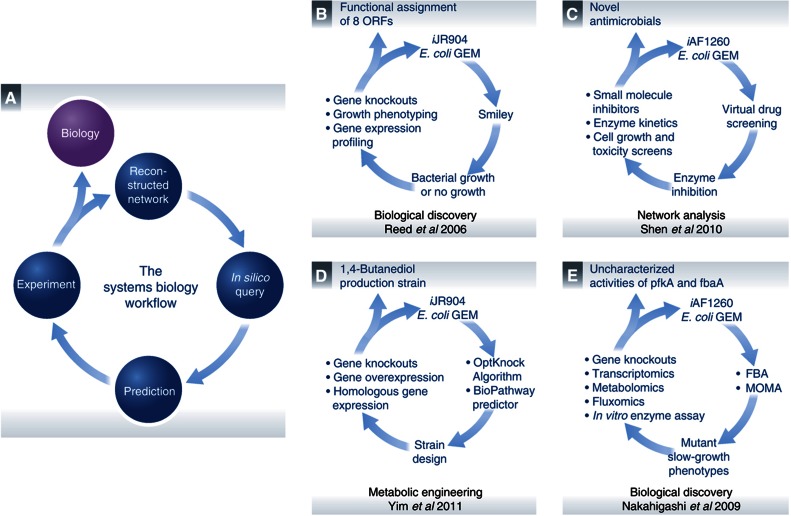
Figure 4.
Iterative workflows. (A) A generic network reconstruction and model-driven systems biology workflow is a cyclic path that iterates between in silico predictions and in vivo observations. This general process has been followed in some of the more influential studies presented in this review. DNA sequencing and bibliomic data can be used to reconstruct and translate a biological system into a mathematical structure. Other omics data types that have been generated can be interpreted in the context of a reconstruction and computational model to analyze organism functions under specific conditions. This information becomes a de facto knowledge base that can be queried through a consortium of analytical methods. The aim of these methods is to hypothesize answers to complex biological questions that can often be nonintuitive or not readily apparent. Experiments can then be designed to test these predictions in order to either confirm GEM-derived explanations or move researchers one iteration closer to the answer. Studies that have successfully iterated through the E. coli GEM workflow that are presented as examples include (B) Reed et al (2006), (C) Shen et al (2010), (D) Yim et al (2011) and (E) Nakahigashi et al (2009).